sgRNA library re-amplification in liquid culture
Erik Haussner, Michael Böttcher
Abstract
In this protocol, we describe a stepwise procedure for the re-amplification of sgRNA libraries in liquid culture. In our hands, this protocol works reliably to amplify pre-cloned sgRNA libraries (e.g. order from Addgene) in a way that preserves the distribution of library elements.
Before start
Keep your original stock safe and aliquoted. For large plasmids with complementary sequences such as LTR sites, keep in mind that repeated reamplification from an already reamplified stock may lead to an accumulation of recombined plasmids and a poorer distribution of library elements.
Steps
Library transformation
Prepare Sample
Thaw
0h 5m 0s on ice.
Pre-cool
Pre-warm 37°C .
Add 100ng Sample into 25µL
Add 25µL of the plasmid/cell mix into a cuvette, electroporate at 1.2 kV, 25 uF and 200 ohm or alternative setting (see note below). Directly after electroporation, add 1mL of pre-warmed
Library recovery
After electroporation, add the 1mL resuspended cells in a 14 ml culture tube and incubate the cells in a thermoshaker 600rpm .
Determination of transformation efficiency
Use a small fraction of your cells to determine the electroporation efficiency of the reamplification.
For 1:10,000 dilution:
Prepare 10µL of recovery culture and dilute in 990µL of 100µL of 1:100 dilution and dilute in 900µL of 100µL on
For 1:1,000,000 dilution:
Take 10µL of the 1:1,000 dilution and dilute in 990µL of
Place the plates in an incubator at 37°C .
Library extraction and quality control
Use rest of recovery to inoculate up to 500mL of 600rpm
On the next day, check for overall coverage via colony counting on
Library preparation and QC
Follow the protocol instructions of the
Determine your final Sample concentration via NanoDrop or Qubit measurement.
Equipment
| Value | Label |
|---|---|
| Qubit 2.0 Fluorometer instrument | BRAND |
| Q33226 | SKU |
| with Qubit RNA HS Assays | SPECIFICATIONS |
Equipment
| Value | Label |
|---|---|
| NanoDrop™ One/OneC Microvolume UV-Vis Spectrophotometer | NAME |
| UV-Vis Spectrophotometer | TYPE |
| Thermo Scientific | BRAND |
| ND-ONE-W | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
Send a sample of your reamplified Sample for Sanger sequencing.
Below we show an example chromatogram of an expected sequencing result. We recommend using sequencing primers 50-100 nt upstream of the sgRNA region. You should see clean traces up- and downstream of the SPACER region, and a noisy 20 nt signal in the SPACER region, due to the sgRNA diversity in your library.
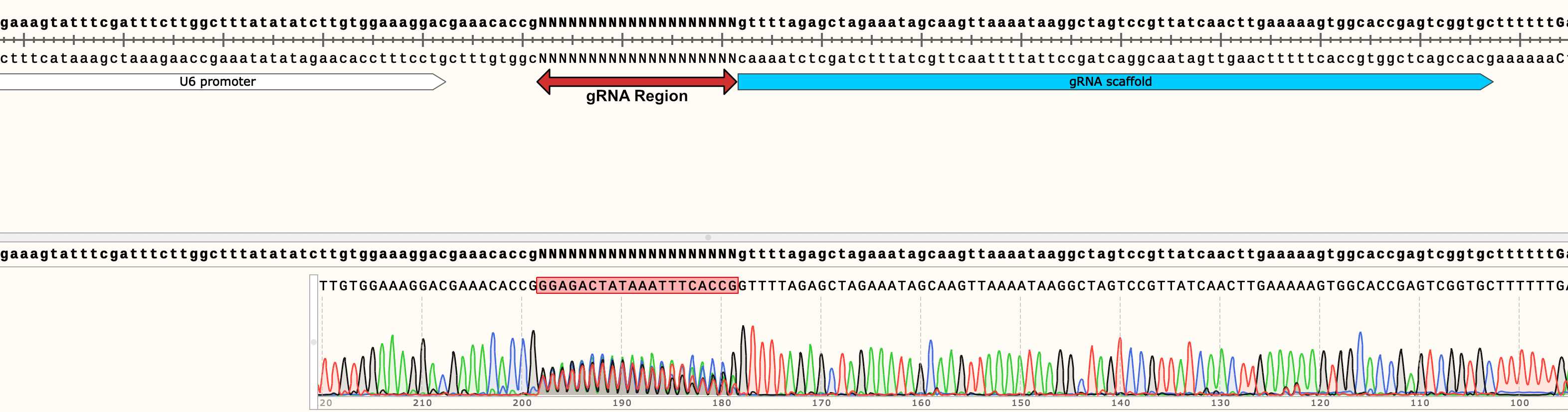
QC: Plasmid recombination check
Since sgRNA library plasmids can recombine during E.coli re-amplification, it is recommended to check for recombination via linearization of 200ng of your reamplified Sample via a restriction digest within the backbone of your library vector.
Prepare a 1g of 100mL
Let the required amount for casting cool down till it is approxametly 50°C and add 1µL of
Pour the warm, still liquid gel into an electroporation chamber and wait until it has cooled down.
Mix your liearized reamplified Sample with
Add your linearized reamplified Sample alongside with the prepared 1h 0m 0s at 120 V.
Check the plasmid size on your gel using UV excitation.

