Smart-seq3xpress
Christoph Ziegenhain, Michael Hagemann-Jensen, Rickard Sandberg
Abstract
Plate-based single-cell RNA-sequencing methods with full-transcript coverage typically excel at sensitivity but are more resource and time-consuming. Here, we miniaturized and streamlined the Smart-seq3 protocol for drastically reduced cost and increased throughput.

Steps
Considerations (PLEASE READ BEFORE START)
• This protocol requires a some type of liquid handler capable of doing nanoliter dispenses. We have tested and used ( Formulatrix Mantis, Dispendix I.Dot & Dispendix I.Dot Mini ). Other (non contact) liquid dispensers should work as well, as long as they can dispense the required volumes accurately.
• All volumes have been scaled to nanoliter volumes, as such the volume your cell is dispensed in matters. We have tested this protocol with an array of FACS machines and cell printers including BD FACSMelody, BD Fusion, BD Influx, Sony SH800S, Cellenion CellenOne, Cytena F.SIGHT Omics, which all typically dispenses the cell in ~5-10nl or less. If your instrument dispenses in higher volumes (> 50nl) , the protocol may either not work or not be as efficient.
• Consider what buffer you use to dispense / sort your single cells in. Since the relative difference between sorted cell volume and lysis volume has overall decreased, common additives like FBS, BSA, EDTA can potentially interfere and affect downstream molecular reaction if present in high enough amounts. As such we recommend if possible to sort in PBS alone, or as recommended by 10x Genomics a solution of PBS + 0.04% BSA at most. Refrain also from using buffers with Mg2+ and Ca2+ or other metal ions for sorting. If EDTA is an absolute must, try and keep the amounts low. Avoid other additives like DNAseI, and Sodium Azide.
• Not all RNase Inhibitors are compatible and some can have severe negative impact on the downstream reaction and ultimately library quality. It is highly recommened to use the RNase Inhibitor used in the protocol (
However quality and efficiency of the protocol might vary with these alternatives. Do NOT use SUPERaseIn RNase Inhibitor (Thermo Fisher Scientific, AM2694) in the general protocol as well as an additive in the final buffer cells are sorted in.
• This protocol in it's current form is not compatible with cell-picking and mouth pipetting for capturing and dispensing single cells.
• Due to the viscosity of the overlays they can compromise the adhesive abilities of PCR seals; especially after storage in -80C. This can be unusual to work with in the beginning. However loose seals have no practical implications, as the method works perfectly well even without seals, or even without heated thermocycler lids during reverse transcription and preamplification PCR. However, heated lid thermocyclers are necessary for the tagmentation reaction, that run independent of overlays. The inert overlay should fully encapsulate the reaction. Furthermore the seal will adhere again better after the initial 72C denaturation step. Even not strictly required, we do recommended to run the method with seals to protect from contamination.
• Take a look at the Guidelines section for more info about Oligos etc. used in this protocol. Recommened storage for oligos are for OligodT30VN and PCR primers and for TSO (The TSO contains RNA bases, so please store it at -80C) -20°C for OligodT30VN and PCR primers and -80°C for TSO (The TSO contains RNA bases, so please store it at -80C)
• Can I change reagent A to reagent B, and oligo X to oligo Y?
• Usually yes it is possible, but user optimization might be required, and results may vary. It is no longer Smart-seq3xpress, and our support will therefore be limited.
• For further details and considerations on method design and decisions please read the supplementary note in the manuscript
Before Starting
This protocol should be carried out in a clean environment. Use ethanol, RNAseZAP, DNA-OFF, or similar to prepare work bench before start.
Work quickly and preferably on ice.
Try and prepare master-mixes right before use, while the plate(s) are finishing the previous incubation step.
Prepare 22% PEG beads for final library clean-up (Optional, but recommended)
This step is optional and can be disregarded if you choose to use
These 22% PEG beads for clean-up is prepared similar to the mcSCRB-seq protocol (mcSCRB-seq)
| A | B |
|---|---|
| Reagent | Amount |
| PEG 8000 | 11g |
| NaCl (5M) | 10mL |
| Tris-HCl (1M, pH 8) | 500uL |
| EDTA (0.5M) | 100uL |
| IGEPAL CA-630 (10% Solution) | 50uL |
| Sodium Azide (10% Solution) | 250uL |
| H2O | Up to 49mL |
| Total | 50mL |
Weight out the PEG8000 in a 50mL Falcon tube, and add all ingredients except IGEPAL CA-630 together. DO NOT add the total amount of water but wait until the PEG is completely dissolved until filling the Falcon tube up to 50mL.
Incubate at 37-40°C and vortex regularly to help dissolve the PEG8000.
Meanwhile the PEG8000 solution is dissolving prepare the Sera-Mag Speed Beads.
Resuspend the bead stock, and pipette 1000µL of bead stock into a 1.5mL tube.
Place on magnet stand and let beads collect. Remove supernatant
Add 1000µL of a 10mM Tris-HCl pH 8, 1mM EDTA (TE) solution and resuspend beads before retuning the tube on the magnet stand.
Remove supernatant and repeat the wash above once more.
Afterwards remove supernatant and add and re suspend the beads in 900µL 10mM Tris-HCl pH 8, 1mM EDTA (TE).
Add the bead solution to the PEG8000 solution above, after it is dissolved.
Add IGEPAL-CA630, and fill up with the remaining H2O to 50mL. Mix it well.
Prepare "overlay" plates
We have tested multiple types of hydrophobic inert overlays such as Silicone Oils, Hydrocarbons, and the commercial Vapor-Lock (Qiagen). They can have different properties, such as viscosities and solidifying temperature. We commenly use Vapor-Lock, Sillicone oil 25 cSt, Silicone Oil 100 cSt (The higher viscosity is better suited for shipping plates).
CAUTION!!! Do not dispense these silicone oils / overlays with your non contact liquid handler. The solutions can "creep" everywhere. Use either manual multichannel pipettes or semi-manual (e.g. Integra ViaFlow) / automatic dispensing (e.g. Agilent Bravo, Tecan Fluent) with tips, and prepare and store in bulk.
Add 3µL of overlay to each well of a 384 well plate. The amount of overlay can be increased if desired.
Quick pulse centrifugation to 1000x g,0h 0m 0s to ensure all is collected in the bottom.
Put on seal and store at Room temperature until use.
Prepare lysis plates
Prepare lysis buffer reaction mix. The mix is prepared for 500 samples. Adjust this to suit your liquid handlers dead volume and pipetting comfort level.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction conc. | uL per reaction | 384 well plate (uL) |
| Triton X100 (10% solution) | 0.1% | 0.003 | 1.5 |
| Poly-ethylene Glycol 8000 (40% solution) | 5% | 0.05 | 25 |
| Spike-ins (optional) | - | - | - |
| RNase Inhibitor (40u/uL) | 0.4u | 0.003 | 1.5 |
| OligodTVN30 (10uM) | 0.125uM | 0.01 | 2.5 |
| dNTPs (10mM / each) | 0.5mM/each | 0.02 | 10 |
| H2O | 0.22 | 109.6 | |
| Total | 0.3uL | 150uL |
Reaction concentrations for PEG8000, OligodT30VN and dNTPs, are adjusted to and reflect their final concentration in the reverse transcription reaction (0.4uL).
• Ensure that PEG is fully mixed into solution, by either pipetting up and down until the liquid is clear, or start with vortexing the required master-mix volume of water and PEG together before adding the remaining reagents.
Add 0.3µL of lysis mix to each well of a 384 well plate containing overlay.
Quick pulse centrifugation to 1000x g,0h 0m 0s to ensure lysis mix is collected in the bottom underneath the overlay before storage until use. For short term storage On ice . For longer term storage, store the lysis plates in a freezer at -20°C or at -80°C if the lysis plates contain Spike-in RNAs.
Sample collection / Cell sorting
Sort single cells into each well containing 0.3µL lysis reaction mix overlayed by 3µL of preferred overlay.
As mentioned in the Considerations section . Be mindful of the solution cells are sorted in, as additives such as BSA, FBS and EDTA, can have negative impact on the downstream molecular reactions.
Seal plates with a cold storage foil seal, do a quick pulse centrifugation to 1000x g,0h 0m 0s and immediately store at -80°C or on dry ice.
Cell lysis
Remove the plate of sorted cells from the -80 freezer and incubate in a thermocycler with heated lid at 72°C for 0h 10m 0s followed by a 4°C hold. Keep the -80C storage seal on for this step, even if it seems loose (read above).
Reverse transcription
While the plate is incubating in step 7, prepare RT reaction mix. The mix is prepared for 500 samples. Adjust this to suit your liquid handlers dead volume and pipetting comfort level.
| A | B | C | D |
|---|---|---|---|
| Reagents | Reaction conc. | uL per reaction | 384 well plate (uL) |
| Tris-HCl pH 8.0-8.4 (1M) | 25mM | 0.01 | 5 |
| NaCl (2.5M) | 30mM | 0.0048 | 2.4 |
| MgCl2 (100mM) | 2.5mM | 0.01 | 5 |
| GTP (100mM) | 1mM | 0.004 | 2 |
| DTT (100mM) | 8mM | 0.03 | 16 |
| RNase Inhibitor (40u/uL) | 0.25u | 0.0025 | 1.25 |
| Smart-seq3xpress TSO (100uM) | 0.75uM | 0.003 | 1.5 |
| Maxima H-minus RT enzyme (200u/uL) | 2u | 0.004 | 2 |
| H2O | 0.03 | 14.9 | |
| Total | 0.1uL | 50uL | |
Add 0.1µL of RT reaction mix into each well, seal the plate with a PCR seal and pulse centrifuge the plate quickly to 1000x g,0h 0m 0s to ensure RT mix merges properly with the lysed cell mix.
Incubate the plate in a thermocycler as following.
| A | B | C |
|---|---|---|
| Temperature | Time | Cycles |
| 42C | 90min | 1x |
| 50C | 2min | 10x |
| 42C | 2min | |
| 85C | 5min | 1x |
| 4C | Hold | - |
Preamplification PCR
While the plate is incubating in step 8, prepare PCR reaction mix. The mix is prepared for 500 samples. Adjust this to suit your liquid handlers dead volume and pipetting comfort level.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction conc. | uL per reaction | 384 well plate (uL) |
| SeqAmp PCR buffer (2x) | 1x | 0.5 | 250 |
| Forward Primer (100uM) | 0.5uM | 0.005 | 2.5 |
| Reverse Primer (100uM) | 0.5uM | 0.005 | 2.5 |
| SeqAmp DNA polymerase (1.25u/uL) | 0.025u | 0.02 | 10 |
| H2O | 0.07 | 35 | |
| Total | 0.6uL | 300uL |
Add 0.6µL of PCR reaction mix to each well seal the plate with PCR seal and pulse centrifuge the plate quickly to 1000x g,0h 0m 0s to ensure RT mix merges properly with the lysed cell mix.
Run the following PCR program in a thermocycler.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial denaturation | 95C | 1min | 1x |
| Denaturation | 98C | 10sec | |
| Annealing | 65C | 30sec | 12-16x* |
| Elongation | 68C | 4min | |
| Final Elongation | 72C | 10min | 1x |
| Hold | 4C | Hold | - |
*To determine the number of PCR cycles it mainly depends on your cells and RNA content. We use as a standard 12 cycles for HEK cells, and 16 cycles for PBMCs.
However, for most cell types a wide range of PCR cycles can be used, yielding similar quality data. Below is a schematic of selected sequencing output metrics from a PCR cycle evaluation run on PBMCs. Each 384 well plate of sorted PBMCs from the same donor, same batch, underwent a different number of PCR cyclers in preampfification. (Take into consideration heterogeneity of PBMCs as each column is a unique 384 well plate of cells)
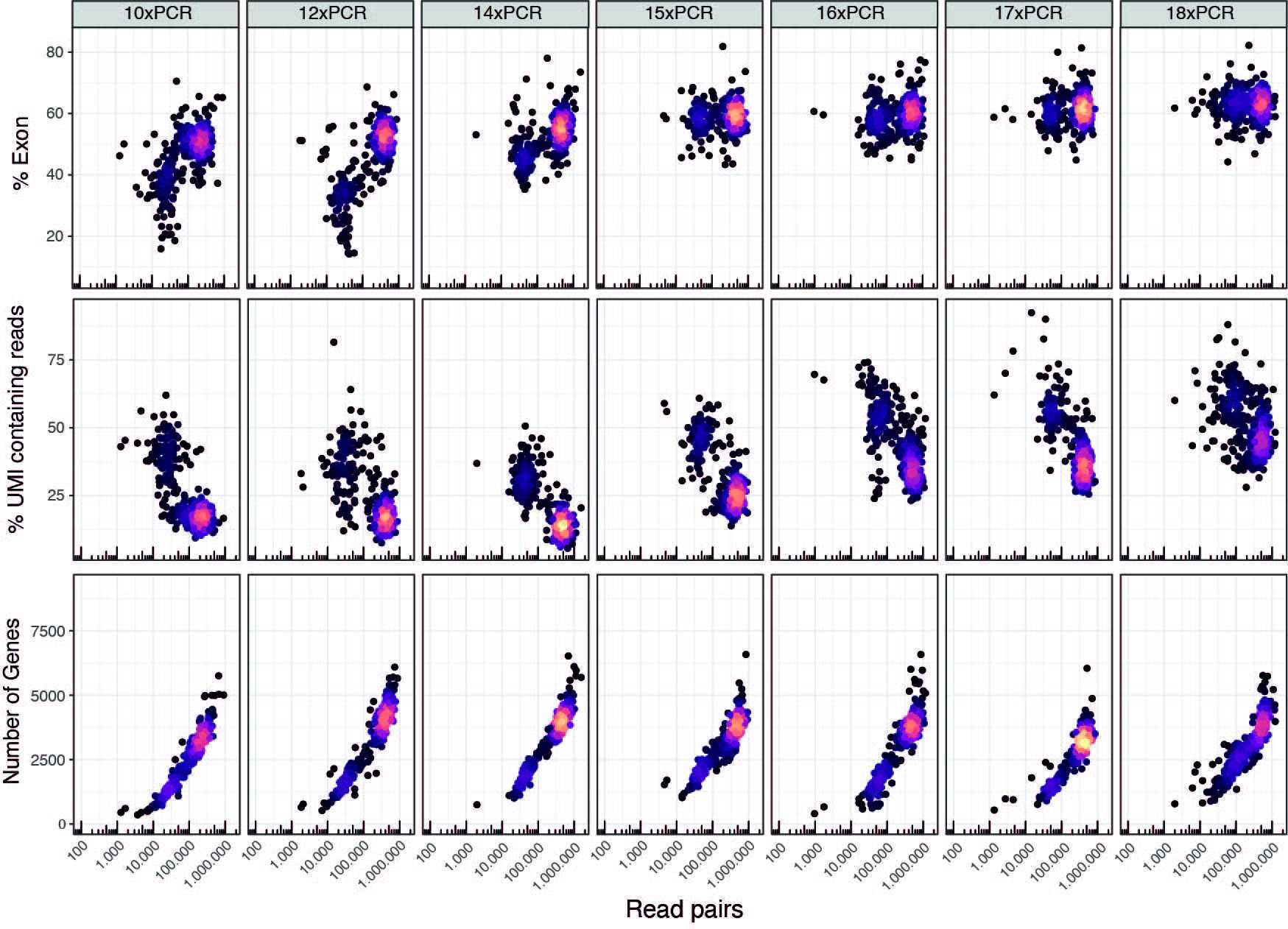
The primary concerns when choosing PCR cycles is
-
To amplify cDNA enough to be well above the genomic DNA levels.
-
Using more cycles means also needing to use more TDE1 Tn5 enzyme per cell to be able to get a good ratio of UMI containing reads to internal reads (~50%). This is not such a massive concern since we use minuscule amounts of TDE1 per cell, If you need to adjust the amount of TDE1 Tn5, we recommend adjustments in the order of +/- 0.0005uL per cell increments.
-
Choosing lower PCR cycles might mean you will need to scale up the amounts of PCR cycles for the tagmentation PCR (see below
Above image in higher quality format.
Dilution
After the preamplification PCR dilute the 1µL cDNA by adding 9µL of
Again, seal the plate and pulse centrifuge to 1000x g,0h 0m 0s to ensure that everything is collected beneath the overlay.
Tagmentation (with index plates)
-
Transfer
1µLof diluted cDNA to a new 384 well plate, and putOn icefor until tagmentation. -
Prepare 4x Tagmentation buffer as following. Aliquots of 4xTD buffer can be stored at
-20°Cfor later use.
| A | B | C |
|---|---|---|
| Reagent | Amount (uL) | Concentration at 4x |
| Tris-HCl pH 7.5 (1M) | 40 | 40mM |
| MgCl2 (100mM) | 200 | 20mM |
| DMF | 200 | 20% |
| H2O | 560 | |
| Total | 1000uL |
- Prepare Tagmentation Mix. Mix is for 500 samples.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction concentration | uL per reaction | 384 well plate (uL) |
| 4x Tagmentation buffer | 1x | 0.5 | 250 |
| TDE1 Tn5 | 0.002* | 1 | |
| H2O | 0.498 | 249 | |
| Total | 1 | 500 |
NB !!! TDE1 Tn5 enzyme comes as a glycerol viscous solution. When pipetting these small uL amounts to your tagmentation mix, be careful to not transfer extra drops that are stuck to the pipette tip. This can cause overtagmentation, which is not bad, but can cause less than expected UMI reads to be captured in sequencing.
*This is a suggestive amount of Tn5, but overall a good starting point that works well with HEK cells (12xPCR preamplification) and PBMCs (16xPCR preamplification). The amount can be changed to meet user specific criteria in terms of ratio of UMI containing reads vs Internal reads, and can be affected by cell-type or amount of preamplification PCR cycles given.
-
Dispense
1µLof Tagmentation Mix to each well of the 384 well plate containing1µLof diluted and pre-dispensed cDNA. Seal and pulse centrifuge the plate down at1000x g,0h 0m 0s. -
Incubate the plate at
55°Cfor0h 10m 0sin a thermocycler. -
To stop the tagmentation reaction and strip off the Tn5 enzyme, add
0.5µLof 0.2% SDS to each well. Seal the plate and quickly centrifuge the plate down before incubation atRoom temperaturefor0h 5m 0s -
Proceed to either substep 11.1 or substep 11.2 depending on your index primer concentration and whether you want to PCR your tagmented libraries in higher or lower volumes. For adding Nextera index primers we recommend preparing index plates with already combined Nextera i5 and i7 indexes at a certain working dilution.
Lower volume indexing PCR, final volume 5uL per well. This can save a bit of cost on Phusion Polymerase
8.1 Add 1µL of premixed index primers.
| A | B | C |
|---|---|---|
| Reagent | Reaction conc. | uL per reaction |
| Premixed custom S50X / N70X index primers (1uM /each) | 0.2uM /each | 1uL |
9.1 Prepare Index PCR mix. Again the calculated amount here for ease is for 500 sample. Please scale this to suit your specific dead volume.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction conc. | uL per reaction | 384 well plate (uL) |
| Phusion HF buffer (5x) | 1x | 1 | 500 |
| dNTPs (10mM/each) | 0.2mM/each | 0.1 | 50 |
| Tween-20 (10%) (*necessary) | 0.025% | 0.0125 | 6.25 |
| Phusion HF Polymerase | 0.01u/uL | 0.025 | 12.5 |
| H2O | 0.3625 | 181.25 | |
| Total | 1.5uL | 750 |
*Addition of Tween-20 to the PCR reaction is necessary to avoid having the remaining SDS affect Phusion DNA polymerase, at the given concentration and amount used in the stop reaction. Tested working amounts of Tween-20 used with Phusion polymerase is 0.005-0.05%. Above 0.05% the reaction starts to get negatively affected by it in our hands. If you change these parameters try and match or be in slight excess with the amount of Tween-20 to the concentration of leftover SDS from the stop solution.
10.1 Add 1.5µL of index PCR mix to each well, seal the plates, apply quick centrifugation to settle everything in the bottom, and incubate in a thermocycler as following:
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Gap filling | 72C | 3min | 1x |
| Initial denaturation | 98C | 30sec | 1x |
| Denaturation | 98C | 10sec | |
| Annealing | 55C | 30sec | 10-14x* |
| Elongation | 72C | 1min | |
| Final elongation | 72C | 5min | 1x |
| Hold | 4C | Hold |
*The amount of PCR cycles needed depends on the starting material, preamplifation cycles given, and amount of tagmentation performed (amount TDE1 Tn5 used per cell). Therefore some user specific optimization may be necessary to suit the needs and wants and final material required.
However as a point of reference:
• HEK cells (12xPCR cycles in preamplification PCR) we use a standard 12xPCR cycles in tagmentation PCR
• PBMCs (10-18xPCR cycles in preamplification PCR) we use 14xPCR cycles in tagmentation PCR.
Standard volume index PCR, final volume 8uL. If you have index plates in the working concentration used for Smartseq3. (0.5micromolar (µM) )
8.2 Add 3.5µL of premixed index primers to each well.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction conc. | uL per. reaction | |
| Premixed custom S50X / N70X index primers (0.5uM/each) | ~0.22uM/each | 3.5uL | |
9.2 Prepare index PCR mix. Again the calculated amount here for ease is for 500 sample. Please scale this to suit your specific dead volume.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction conc. | uL per reaction | 384 well plate (uL) |
| Phusion HF buffer (5x) | 1x | 1.6 | 800 |
| dNTPs (10mM/each) | 0.2mM/each | 0.16 | 80 |
| Tween-20 (10%) (optional*) | 0.01% | 0.008 | 4 |
| Phusion HF Polymerase | 0.01u/uL | 0.04 | 20 |
| H2O | 0.192 | 96 | |
| Total | 2uL | 1000uL |
*Adding a bit of Tween-20 to the PCR reaction can help protect the DNA polymerase a bit against SDS from previous stop reaction. This trick can also help other polymerases to work better like KAPA, Vent etc.
10.2 Add 2µL of index PCR mix to each well, seal the plates, apply quick centrifugation to settle everything in the bottom, and incubate in a thermocycler as following:
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Gap filling | 72C | 3min | 1x |
| Initial denaturation | 98C | 30sec | 1x |
| Denaturation | 98C | 10sec | |
| Annealing | 55C | 30sec | 10-14x* |
| Elongation | 72C | 1min | |
| Final Elongation | 72C | 5min | 1x |
| Hold | 4C | Hold | - |
*The amount of PCR cycles needed depends on the starting material, preamplification cycles given, and amount of tagmentation performed (TDE1 Tn5 amount added per cell). As such it might require a bit of user optimization, depending on wants, needs, and final material needed.
However as a reference point:
• HEK cells (12xPCR cycles in preamplification PCR) we use as standard 12xPCR cycles in tagmentation PCR. (10xPCR is confirmed working as well)
• PBMCs (10x-18xPCR cycles preamplification PCR) we use 14xPCR cycles in tagmentation PCR.
Tagmentation (Dessicated index primer startegy)
To save even further on general plastic consumption and tip use, we designed a way of tagmention & indexing in which one prepares predispensed "tagmentation" plates containing premixed dessicated indexes. Hence you can with the same set of tips prepare multiple tagmentation plates containing the same indexes for storage. Of course it is very important to note that you need to change tips when/if you change premixed source indexes so as to not mix/contaminate your premixed stock indexes with different set of indexes.
This method is the most cost-effective implementation of Smart-seq3xpress and especially suited for large-scale projects or preparing large amounts of plates in advance.
Depending on your premixed index plate concentrations, dispense xµL of indexes aiming for a concentration of between 0.2micromolar (µM) -0.5micromolar (µM) for a 5uL final volume reaction. It can also be performed in a 7.5uL final volume reaction, simply adjust the concentration accordingly to fit the higher volume
Examples for a 5uL final reaction volume
For 0.5micromolar (µM) premixed indices add 2.5µL to each well of a 384 well PCR plate (results in0.25micromolar (µM)) .
For 1.0micromolar (µM) premixed indicies add 1.25µL to each well of a 384 well PCR plate. (0.25micromolar (µM) ).
Put the PCR plates onto an open thermocycler and incubate at 95°C without seal for 0h 1m 0s - 0h 5m 0s depending on the amount if index primers added to each well.
In the process or after visually inspect that all the liquid has evaporated from the wells.
Seal and store the plates at -20°C until use.
-
Transfer
1µLof prediluted cDNA from step 10 into a 384 well PCR plate containing dessicated index primers. -
Prepare tagmentation mix (see Step 11 for the recipe for 4xTagmentation buffer). Shown mix is for 500 samples.
| A | B | C | D |
|---|---|---|---|
| Reagent | Reaction concentration | uL per reaction | 384 well plate (uL) |
| 4x tagmentation buffer | 1x | 0.5 | 250 |
| TDE1 Tn5 | 0.002* | 1 | |
| H2O | 0.498 | 249 | |
| Total | 1uL | 500uL |
NB !!! TDE1 Tn5 enzyme comes as a glycerol viscous solution. When pipetting these small uL amounts to your tagmentation mix, be careful to not transfer extra drops that are stuck to the pipette tip. This can cause overtagmentation, which is not bad, but can cause less than expected UMI reads to be captured in sequencing.
*This is a suggestive amount of Tn5, but overall a good starting point that works well with HEK cells (12xPCR preamplification) and PBMCs (16xPCR preamplification). The amount can be changed to meet user specific criteria in terms of ratio of UMI containing reads vs Internal reads, and can be affected by cell-type, amount of preamplification PCR cycles given.
-
Dispense
1µLof Tagmentation Mix to each well of the 384 well plate containing1µLof cDNA. Seal and pulse centrifuge the plate down at1000x g,0h 0m 0s. -
Incubate the plate at
55°Cfor0h 10m 0sin a thermocycler. -
To stop the tagmentation reaction and strip off the Tn5 enzyme, add
0.5µLof 0.2% SDS to each well. Seal the plate and quickly centrifuge the plate down before incubation atRoom temperaturefor0h 5m 0s -
Prepare Index PCR mix. Again the calculated amount here for ease is for 500 sample. Please scale this to suit your specific dead volume.
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| Reagent | Reaction concentration | uL per reaction (5uL final) | 384 well plate (uL/5uL final) | uL per reaction (7.5uL final) | 384 well plate (uL/7.5uL final) |
| Phusion HF buffer (5x) | 1x | 1 | 500 | 1.5 | 750 |
| dNTPs (10mM/each) | 0.2mM/each | 0.1 | 50 | 0.15 | 75 |
| Tween-20 (10%) | 0.025% / 0.01% | 0.0125 | 6.25 | 0.0075 | 3.75 |
| Phusion DNA polymerase (2u/uL) | 0.01u/uL | 0.025 | 12.5 | 0.0375 | 18.75 |
| H2O | 1.3625 | 681.25 | 3.305 | 1652.5 | |
| Total | 2.5uL | 1250uL | 5uL | 2500uL |
7 Add either 2.5µL or 5µL of index PCR mix to each well depending on chosen final volume, seal the plates, apply quick centrifugation to settle everything in the bottom, and incubate in a thermocycler as following:
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Gap filling | 72C | 3min | 1x |
| Initial denaturation | 98C | 30sec | 1x |
| Denaturation | 98C | 10sec | |
| Annealing | 55C | 30sec | 10-14x* |
| Elongation | 72C | 1min | |
| Final elongation | 72C | 5min | 1x |
| Hold | 4C | Hold | - |
*The amount of PCR cycles needed depends on the starting material, preamplifation cycles given, and amount of tagmentation performed (amount TDE1 Tn5 used per cell). Therefore some user specific optimization might be necessary to suit the needs and wants and final material required.
However as a point of reference:
• HEK cells (12xPCR cycles in preamplification PCR) we use a standard 12xPCR cycles in tagmentation PCR
• PBMCs (10-18xPCR cycles in preamplification PCR) we use 14xPCR cycles in tagmentation PCR.
Library pooling by spin-out and bead clean-up
To quickly pool a plate or multiple plates together we designed a centrifugaton holder heavily inspired from Quartz-seq2, although more accessible. This holder can easily be 3D printed, and fits together with

Put the holder around the reservoir, and add onto the PCR plate containing the final tagmented libraries upside down. Liquid should stay in the wells, by surface tension until centrifugation, but again do this with a slight amount of caution. See pictures if in doubt.


Once this is assembled, put in a centrifuge and pulse to max 100x g,0h 0m 0s .

Collect the pooled library in a fitting tube (depending on how many plates you choose to spin in the same reservoir) and purify the library with 22% PEG Clean-up beads or similar at a ratio of 0.7 : 1 beads to sample .
Mix the beads and sample gently by pipetting up and down and incubate for 0h 8m 0s at Room temperature
Place on magnet and let beads settle for roughly 0h 5m 0s to 0h 10m 0s before discarding the supernatant
Wash twice with freshly made 80% Ethanol
Remove Ethanol and let the bead pellet air dry for at least 0h 5m 0s (Until the pellet is no longer shiny)
Elute the beads in 40µL of UltraPure Water, resuspend beads and incubate for 0h 5m 0s
Final Library QC
Use Qubit fluorometer (Qubit dsDNA HS Assay) or similar to quantify the final library pool / pools.
Run the final library on an Agilent Bioanalyzer (High Sensitivity DNA chip) to inspect quality and get median base-pair length information.
Sequencing
The sequencing ready library should be sequenced on any MGI or Illumina compatible sequencer, either Single-end or Paired-end, depending on the question and need.
If sequencing on a MGI sequencer convert the final library into MGI compatible single stranded circles utililizing the Universal Library Conversion Kit (App-A). Follow the user manual.
| A | B | C | D | E |
|---|---|---|---|---|
| Oligo Name | Vendor | Purification | Stock Concentration | Sequence |
| Splint-Oligo (App-A) | IDT | HPLC | 100uM | TCGCCGTATCATTCAAGCAGAAGACG |
| MDA Primer | IDT | HPLC | 100uM | CGTATGCCGTCTTCTGCTTGAATGATACGGCGAC |
| Read1 Sequencing Primer | IDT | HPLC | 100uM | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG |
| Read2 Sequencing Primer | IDT | HPLC | 100uM | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG |
| I5-index primer | IDT | HPLC | 100uM | CTGTCTCTTATACACATCTGACGCTGCCGACGA |
| I7-index primer | IDT | HPLC | 100uM | CCGTATCATTCAAGCAGAAGACGGCATACGAGAT |
Table of Sequencing primers and oligos needed for MGI sequencing of Nextera Style libraries.
Data processing
For primary data processing we highly recommend using the zUMIs pipeline. There are many specifics and features in the pipeline that are directly geared towards preparing and preprocessing Smart-seq3 data properly.
Software
| Value | Label |
|---|---|
| zUMIs | NAME |
| Linux | OS_NAME |
| https://github.com/sdparekh/zUMIs | REPOSITORY |
| https://www.biorxiv.org/content/early/2017/10/18/153940 | LINK |
For data sequenced on Illumina sequencers:
When sequencing has completed convert the binary base-call files (BCL) to fastq files.
Use bcl2fastq (bcl2fastq v2.20).
Software
| Value | Label |
|---|---|
| bcl2fastq | NAME |
| Illumina | DEVELOPER |
| https://emea.support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html | LINK |
| 2.20 | VERSION |
Fastq files for zUMIs should be processed without demultiplexing in the following manner, instead of demultiplexing per cell.
#Example of preparing data from a 150bp paired-end sequencing run
bcl2fastq --use-bases-mask Y150N,I8,I8,Y150N --no-lane-splitting --create-fastq-for-index-reads -R /mnt/storage1/NextSeqNAS/191011_NB502120_0154_AHVG7JBGXB
Remove the flag --no-lane-splitting if the cell barcodes or index primers have been reused on the different lanes.
Fastq files are now ready and compatible with the zUMIs pipeline. An example of how config .yaml files should look like for zUMIs can be seen below.
#Smartseq3xpress.yaml (Illumina Paired-end 150bp)
project: Smartseq3xpress
sequence_files:
file1:
name: /Smartseq3xpress/fastq_files/Read1.fastq.gz
base_definition:
- cDNA(25-150)
- UMI(12-21)
find_pattern: ATTGCGCAATG;2
file2:
name: /Smartseq3xpress/fastq_files/Read2.fastq.gz
base_definition:
- cDNA(1-150)
file3:
name: /Smartseq3xpress/fastq_files/Index1.fastq.gz
- BC(1-10)
file4:
name: /Smartseq3xpress/fastq_files/Index2.fastq.gz
- BC(1-10)
reference:
STAR_index: /genomes/Human/STAR7idx_noGTF/
GTF_file: /genomes/Human/Homo_sapiens.GRCh38.95.chr.gtf
additional_STAR_params: '--clip3pAdapterSeq CTGTCTCTTATACACATCT'
additional_files:
out_dir: /Smartseq3xpress/zUMIs
num_threads: 20
mem_limit: 50
filter_cutoffs:
BC_filter:
num_bases: 4
phred: 20
UMI_filter:
num_bases: 3
phred: 20
barcodes:
barcode_num: ~
barcode_file: /Smartseq3xpress/expected_barcodes.txt
automatic: no
BarcodeBinning: 1
demultiplex: no
nReadsperCell: 100
discardReads: yes
counting_opts:
introns: yes
downsampling: '0'
strand: 1
Ham_Dist: 1
velocyto: no
primaryHit: yes
twoPass: no
make_stats: yes
which_Stage: Filtering
samtools_exec: samtools
pigz_exec: pigz
STAR_exec: STAR
Rscript_exec: Rscript
For data sequenced on a MGI sequencer:
Fastq files should be available after completed sequencing run, in the following format, that is directly compatible with zUMIs pipeline; Read_1.fastq, Read_2.fastq. The index barcodes are located as the last bases of read2.
#Smartseq3xpress.yaml (MGI, Paired-end 150bp)
project: Smartseq3xpress
sequence_files:
file1:
name: /Smartseq3xpress/fastq_files/Read1.fastq.gz
base_definition:
- cDNA(25-150)
- UMI(12-21)
find_pattern: ATTGCGCAATG;2
file2:
name: /Smartseq3xpress/fastq_files/Read2.fastq.gz
base_definition:
- cDNA(1-150)
- BC(151-170)
reference:
STAR_index: /genomes/Human/STAR7idx_noGTF/
GTF_file: /genomes/Human/Homo_sapiens.GRCh38.95.chr.gtf
additional_STAR_params: '--clip3pAdapterSeq CTGTCTCTTATACACATCT'
additional_files:
out_dir: /Smartseq3xpress/zUMIs
num_threads: 20
mem_limit: 50
filter_cutoffs:
BC_filter:
num_bases: 4
phred: 20
UMI_filter:
num_bases: 3
phred: 20
barcodes:
barcode_num: ~
barcode_file: /Smartseq3xpress/expected_barcodes.txt
automatic: no
BarcodeBinning: 1
demultiplex: no
nReadsperCell: 100
discardReads: yes
counting_opts:
introns: yes
downsampling: '0'
strand: 1
Ham_Dist: 1
velocyto: no
primaryHit: yes
twoPass: no
make_stats: yes
which_Stage: Filtering
samtools_exec: samtools
pigz_exec: pigz
STAR_exec: STAR
Rscript_exec: Rscript
FOR MORE INFO:
Preprocessing (Basic look at zUMis output)
To get a quick look and generate a small stats file with the most common metrics from the zUMIs output data, one can do as following in R.
#Example of how to quickly generate an overview of zUMIs output data in R
library(data.table)
dge <- readRDS("/Smartseq3xpress/zUMIs/zUMIs_output/expression/Smartseq3xpress.dgecounts.rds")
stats <- fread("/Smartseq3xpress/zUMIs/zUMIs_output/stats/Smartseq3xpress.readspercell.txt")[!RG %in% "bad"]
stats <- dcast(stats, RG~type, value.var = "N")
stats[, nreadpairs := Ambiguity+Exon+Intergenic+Intron+Unmapped]
reads <- fread("/Smartseq3xpress/zUMIs/zUMIs_output/Smartseq3xpresskept_barcodes_binned.txt.BCUMIstats.txt")
reads[, UMIfraction := nUMItag/(nNontagged+nUMItag)]
stats <- merge(stats,reads, by.x="RG", by.y="XC")
exonic_UMIs <- as.matrix(dge$umicount$exon$all)
exonic_Reads <- as.matrix(dge$readcount$exon$all)
complexity <- data.table( RG = colnames(exonic_UMIs),
nGenes = colSums(exonic_Reads>0),
nGenesUMIs = colSums(exonic_UMIs>0),
nUMIs = colSums(exonic_UMIs))
stats <- merge(stats,complexity, by = "RG")
stats[,pct_coding := ((Exon+Intron)/nreadpairs)*100]





