Improving Species Level-taxonomic Assignment from 16S rRNA Sequencing Technologies
David Bars-Cortina, David Bars-Cortina, Ferran Moratalla-Navarro, Ferran Moratalla-Navarro, Ainhoa García-Serrano, Ainhoa García-Serrano, Núria Mach, Núria Mach, Lois Riobó-Mayo, Lois Riobó-Mayo, Jordi Vea-Barbany, Jordi Vea-Barbany, Blanca Rius-Sansalvador, Blanca Rius-Sansalvador, Silvia Murcia, Silvia Murcia, Mireia Obón-Santacana, Mireia Obón-Santacana, Victor Moreno, Victor Moreno
Abstract
Analysis of the bacterial community from a 16S rRNA gene sequencing technologies requires comparing the reads to a reference database. The challenging task involved in annotation relies on the currently available tools and 16S rRNA databases: SILVA, Greengenes and RDP. A successful annotation depends on the quality of the database. For instance, Greengenes and RDP have not been updated since 2013 and 2016, respectively. In addition, the nature of 16S sequencing technologies (short reads) focuses mainly on the V3-V4 hypervariable region sequencing and hinders the species assignment, in contrast to whole shotgun metagenome sequencing.
Here, we combine the results of three standard protocols for 16S rRNA amplicon annotation that utilize homology-based methods, and we propose a new re-annotation strategy to enlarge the percentage of amplicon sequence variants (ASV) classified up to the species level. Following the pattern (reference) method: DADA2 pipeline and SILVA v.138.1 reference database classification (Basic Protocol 1), our method maps the ASV sequences to custom nucleotide BLAST with the SILVA v.138.1 (Basic Protocol 2), and to the 16S database of Bacteria and Archaea of NCBI RefSeq Targeted Loci Project databases (Basic Protocol 3).
This new re-annotation workflow was tested in 16S rRNA amplicon data from 156 human fecal samples. The proposed new strategy achieved an increase of nearly eight times the proportion of ASV classified at the species level in contrast to the reference method for the database used in the present research. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Sample inference and taxonomic profiling through DADA2 algorithm.
Basic Protocol 2 : Custom BLASTN database creation and ASV taxonomical assignment.
Basic Protocol 3 : ASV taxonomical assignment using NCBI RefSeq Targeted Loci Project database.
Basic Protocol 4 : Definitive selection of lineages among the three methods.
INTRODUCTION
The 16S ribosomal RNA (rRNA) gene sequencing is widely used for microbiota analysis with next-generation sequencing (NGS) technologies. The 16S rRNA gene has nine hypervariable regions (V1 to V9, Chakravorty et al., 2007), but only the V3 and V4 regions are commonly targeted for short-read sequencing and microbial community analysis. These regions have been found to balance sequencing diversity and technical challenges associated with their analysis (López-Aladid et al., 2023). However, the taxonomical annotation when using these regions is often restricted to the genus level because species resolution cannot be achieved (Gwak and Rho, 2020; Hiergeist et al., 2023).
Analysis of the bacterial community from a 16S rRNA amplicon data includes comparing the reads to a reference database. A successful annotation depends on the raw data and database reference quality. To process amplicon sequencing data from raw reads to taxa abundance tables, several bioinformatic pipelines have been developed, such as Quantitative Insights into Microbial Ecology 2 (QIIME2, Bolyen et al., 2019), DADA2 (Callahan et al., 2016), and mothur (Schloss et al., 2009). All these abovementioned pipelines involve mapping reads to taxonomical reference databases. Three common standard 16S rRNA databases are SILVA (Quast et al., 2013), Greengenes (McDonald et al., 2012), and RDP (Wang et al., 2007). In contrast to SILVA, Greengenes and RDP have not been updated since 2013 and 2016, respectively. These databases also differ significantly in their size, content, and how they are curated.
Moreover, different from whole shotgun metagenome sequencing, which analyzes the collective genetic material of all microorganisms in a particular sample, 16S rRNA sequencing presents low species taxa discrimination capacity because of this gene homology between some species (Gwak & Rho, 2020). Strategies are warranted to enhance the taxonomical classification of 16S RNA sequencing data and obtain more accurate insights into the microbial composition of samples. Therefore, here we used a modified custom in-house developed R code to improve the taxonomical classification of the amplicon sequence variants (ASV) obtained from complex communities in fecal samples. Following the default taxonomy assignment obtained from SILVA v.138.1 databases through DADA2 functions assignTaxonomy and addSpecies , the chimera-free ASVs were submitted towards a custom BLASTN database constructed from the SILVA v.138.1 databases, using a robust E-value of 1e-50 and identity threshold of 99.5%. Furthermore, in parallel, the same chimera-free ASV were mapped to the NCBI RefSeq Targeted Loci Project Archaea and Bacteria database (https://www.ncbi.nlm.nih.gov/refseq/targetedloci/) using Kraken2 (Wood et al., 2019) and Bracken 2 (Lu et al., 2017) as a metagenomic classifier.
Each ASV classification method was checked for similarities and discrepancies through an R code automatic checkout to establish the best ASV taxonomic level of resolution. This gave additional confidence about the taxonomic lineages obtained.
STRATEGIC PLANNING
Hardware
The present protocol can be run on Linux, MacOS, or Windows (with Subsystem for Linux) operating system, with sufficient available random-access memory (RAM) and disk space. The most resource-intensive software in terms of computing requirements are DADA2 and Kraken2 (NCBI RefSeq Targeted Loci Project database). As a general recommendation, it is advisable to ensure a minimum of 32 GB of RAM and at least approximately 140 GB of disk space. Therefore, depending on the number of samples to be analyzed, working on a multicore CPU system or a high-performance computing system is highly encouraged.
Flowchart
The flowchart (Fig. 1) shows the steps in the proposed protocol to offer a general idea of the processes described below. This workflow can be considered as a graphic summary of the process. In contrast to Basic Protocol 1, Basic Protocols 2 and 3 show a drop in the output raw ASV numbers possibly due to the stringent homology required to define a match for the blast algorithm but not for the E-value (four more raw ASV classified when increasing to an E-value of 1e—10) and the different taxonomical reference database used in Basic Protocol 3 (NCBI RefSeq Targeted Loci Project). Nevertheless, after the final merge of all the information (Basic Protocol 4), the curated ASV (without non-human gut, Mitochondria, and Chloroplast lineages) grows up and gives additional confidence about the taxonomic lineages obtained.
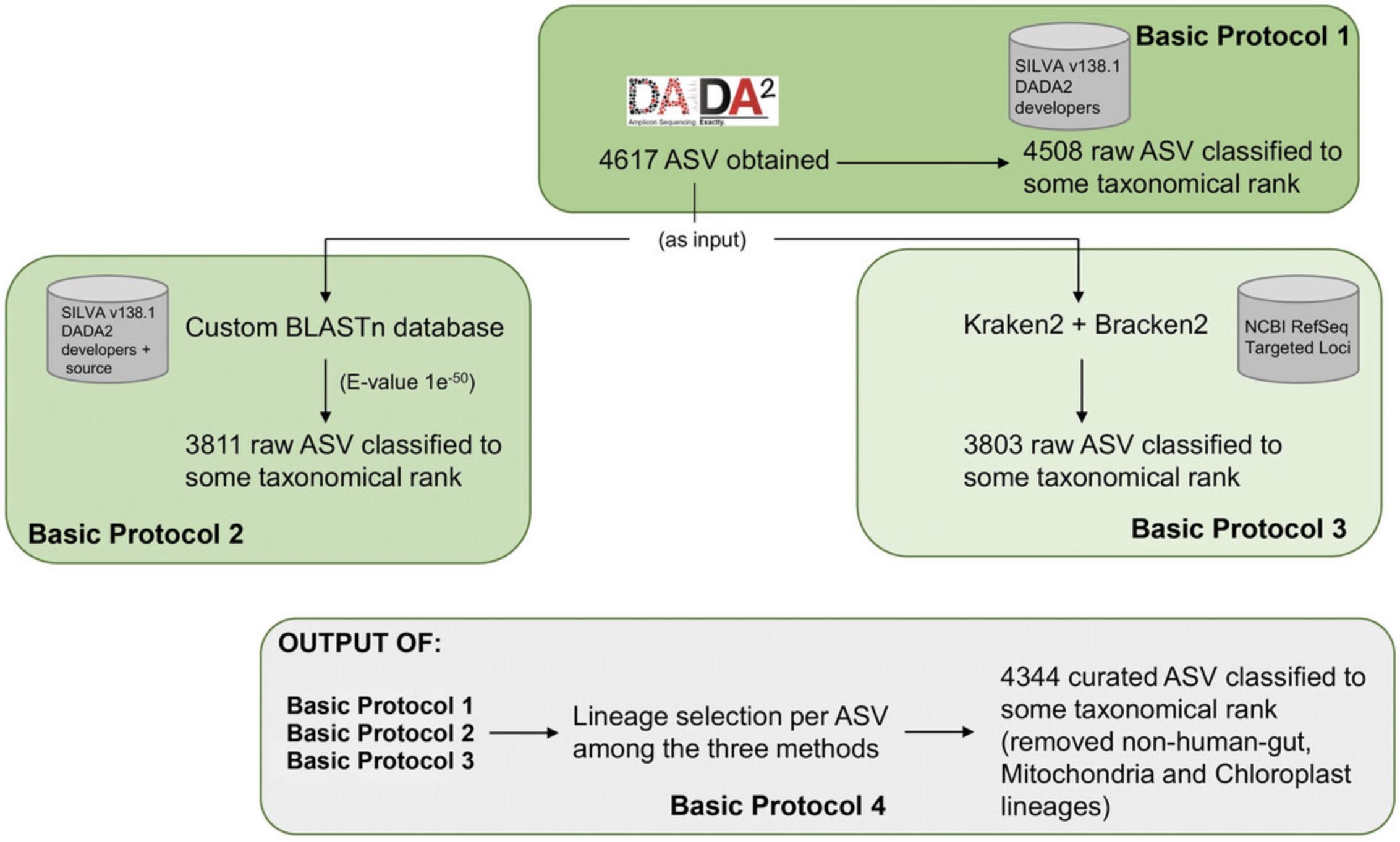
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: SAMPLE INFERENCE AND TAXONOMIC PROFILING THROUGH DADA2 ALGORITHM
This protocol presents the results obtained from DADA2 pipeline to use the ASV sequences, which is the input data for the other protocols described here. The DADA2 pipeline is extensively detailed in the DADA2 developer's webpage (https://benjjneb.github.io/dada2/tutorial.html) ; therefore, we only state the steps followed. Briefly, low-quality reads were filtered and trimmed out based on the observed quality profiles by using the filterAndTrim function, truncating forward and reverse reads below 290 and 230, respectively, and considering a value of 2 as the maximum expected error. In detail, the function arguments for filterAndTrim are:
maxN=0, maxEE=c(2,2), truncQ=2, trimLeft=10, truncLen=c(290.230), rm.phix=T, compress=T, multithread = T (the last argument is indicating the use of multiple cores)
Furthermore, the first 10 nucleotides of each read were removed. We combined identical sequencing reads into unique sequences, made a sample inference from the matrix of estimated learning errors, and merged paired reads. For the sample inference step (see https://benjjneb.github.io/dada2/tutorial.html) the argument of the pool was defined as True. Chimeras and contaminants are often rare but spread across samples, making them much more effectively-identified when the samples are pooled (pool =T). Chimeric sequences were removed by using the removeBimeraDenovo function and taxonomy was assigned utilizing the SILVA 16S rRNA database (v.138.1).
Necessary Resources
Hardware
- Linux, MacOS, or Windows (with Subsystem for Linux) operating system, with sufficient available random-access memory (RAM) and disk space (see Strategic Planning)
Software
- The following software must be installed and available in the PATH environment variable to be executable as a binary system:
- R (v4.1.2): (https://cran.r-project.org/bin/windows/base/old/4.1.2/)
- Rstudio (v1.4.1106): (https://posit.co/download/rstudio-desktop/)
- We recommend having a fundamental understanding of R and RStudio, along with familiarity with their basic commands, which will be utilized in the current protocol.
- R packages:
- DADA2 (v1.22): (https://benjjneb.github.io/dada2/dada-installation.html)
- DECIPHER (v2.22.0): (https://bioconductor.org/packages/release/bioc/html/DECIPHER.html)
- ggplot2 (v3.4.1): (https://cran.r-project.org/web/packages/ggplot2/index.html)
- phangorn (v2.6.2): (https://cran.r-project.org/web/packages/phangorn/index.html)
- tidyverse (v1.3.1): (https://cran.r-project.org/web/packages/tidyverse/index.html)
- R script files can be run interactively in R/Rstudio or in command line with Rscript. We will use Rscript in this protocol.
Files
- seqtab.nochim_pooling.rds, ASV table file obtained after chimeras removal (see https://benjjneb.github.io/dada2/tutorial.html).
Sample files
- All the required files of the present protocol are included in the Figshare link (see Data availability statement) to show the protocol process.
1.Download the sample and reference files.
- wget https://zenodo.org/record/4587955/files/silva_nr99_v138.1_train_set.fa.gz
- wget https://zenodo.org/record/4587955/files/silva_species_assignment_v138.1.fa.gz
2.Assignment of the taxonomy as follows:
-
Once the ASV table without chimeras (fileseqtab.nochim_pooling.rds, Table1) is obtained, the DNA sequence assigned for each ASV is mapped to a specific Bacteria or Archaea lineage.
-
Open, customize the file 08.assign_taxonomy.R to your directory pathway and run:
Rscript 08.assign_taxonomy.R. As shown in the R script file, you need the object seqtab.nochim_pooling.rds. Submit this object first to assignTaxonomy and second to addSpecies functions, pay attention to the R script of the reference databases that use each of both functions.
| ASV DNA sequence 1 | ASV DNA sequence n | |
|---|---|---|
| Sample 1 | Reads 1,1 | Reads 1,n |
| Sample m | Reads m,1 | Reads m,n |
3.Phylogenetic tree (optional step). To create a phylogenetic tree, if you consider opportune to use in your 16S rRNA downstream statistical analysis, customize to your directory pathway and run:
Rscript 09.phylogenetic_tree.R
To perform this step, apart from the dada2 R package, you also need the R packages of phangorn and DECIPHER.
This script will generate output, which we will use downstream: tree_dada2_16S.rds
4.Obtention of the final rds objects: Run Rscript 10.object_rds.R
Basic Protocol 2: CUSTOM BLASTN DATABASE CREATION AND ASV TAXONOMICAL ASSIGNMENT
This protocol includes two steps. The first one describes the basic bash and R script commands to follow for creating a custom BLASTN database that will be used to classify the ASV sequences obtained in Basic Protocol 1.The second step describes the procedure from the BLASTN output to obtain a specific lineage based on E-value and percentage of identical matches (pident) parameters from the BLAST tool and other user-defined parameters detailed on an R script. All this process has been automatized to be robust and reproducible over time. Nevertheless, the output obtained in this protocol needs manual checks (but only for cases where only one specific lineage could be defined for a particular ASV sequence).
Necessary Resources
Hardware
- Linux, MacOS or Windows (with Subsystem for Linux) operating system, with sufficient available random-access memory (RAM) and disk space (see Strategic Planning)
Software
- The following software must be installed and available in the PATH environment variable to be executable as a binary system:
- BLAST (v2.7.1): (https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.7.1/)
- Python (v3.9.12): (https://www.python.org/downloads/)
- R (v4.1.2): (https://cran.r-project.org/bin/windows/base/old/4.1.2/)
- Rstudio (v1.4.1106): (https://posit.co/download/rstudio-desktop/)
- SeqKit (v2.3.0): (https://github.com/shenwei356/seqkit)
- Linux system commands: wget, gzip, tr, awk, grep, nl, paste
- We recommend having a fundamental understanding of bash commands, and a minimum reading of the BLASTN and SeqKit basic user manual is recommended.
Files
- All the input files are detailed in the present protocol and available on the Figshare link.
Sample files
- See Basic Protocol 2 on the Figshare link.
1.Download databases to create custom BLASTN database.
In a specific directory of your choice, download the following reference SILVA databases (some used in Basic Protocol 1). Nevertheless, we use another species multifasta file not used in the DADA2 standard pipeline in the addSpecies R function (see Basic Protocol 1). Furthermore, we also used the original SILVA 138 reference database instead of the DADA2-trained reference customized by DADA2 developers.
- wget https://www.arb-silva.de/fileadmin/silva_databases/release_138.1/Exports/SILVA_138.1_SSURef_NR99_tax_silva.fasta.gz
- wget https://zenodo.org/record/4587955#.ZEaxi2j7SUk/silva_species_assignment_v138.1.fa.gz
- wget https://zenodo.org/record/4587955#.ZEaxi2j7SUk/silva_nr99_v138.1_wSpecies_train_set.fa.gz
2.Decompress all three databases.
- gzip -d SILVA_138.1_SSURef_NR99_tax_silva.fasta.gz
- gzip -d silva_nr99_v138.1_wSpecies_train_set.fa.gz
- gzip -d silva_species_assignment_v138.1.fa.gz
3.Change .fasta extension to .fa:
- mv SILVA_138.1_SSURef_NR99_tax_silva.fasta SILVA_138.1_SSURef_NR99_tax_silva.fa
4.Concatenate all three fasta files:
- cat *.fa >> silva_dada2_arb.fa
Check if repeated identifiers exist (use check_no_duplicates.py provided in the Figshare link or download from the GitHub source: https://github.com/peterjc/galaxy_blast/blob/master/tools/ncbi_blast_plus/check_no_duplicates.py):
- python3 check_no_duplicates.py silva_dada2_arb.fa
- 4.1 Error obtained:
- BLAST Database creation error: Error: Duplicate seq_ids are found:
- LCL|BACTERIA;PROTEOBACTERIA;GAMMAPROTEOBACTERIA;PSEUDOMONADALES;PSEUDOMONADACEAE;PSEUDOMONAS;
5.Create a new identifier for all the sequences in the multifasta file of silva_dada2_arb.fa (solve 4.1):
- cat silva_dada2_arb.fa | seqkit replace -p.+ -r “{nr}” --nr-width 7 > 3basesdades.fa
6.Create a new directory where you will create the custom BLASTN database.
- mkdir database
Move the 3basesdades.fa (that contains no repeated identifiers) to the database directory:
- mv 3basesdades.fa./database
7.Run BLASTN in your machine.
- makeblastdb -in 3basesdades.fa -parse_seqids -title silva138_1_dada2 -dbtype nucl -max_file_sz ‘2GB’ -out customblastdatabase
- Once run, check in the database directory that you have the newly created files:
- customblastdatabase.nin, customblastdatabase.nhr, customblastdatabase.nsq, customblastdatabase.nsi, customblastdatabase.nsi, customblastdatabase.nsd and customblastdatabase.nog
The custom BLASTN database is created satisfactorily.
8.Create another txt file from the BLASTN database.
- Locate the folder that harbors the silva_dada2_arb.fa file and perform the next bash commands:
- grep -e “>” silva_dada2_arb.fa > headers.txt
- nl -nrz headers.txt > headers_2.txt
- cat headers_2.txt | cut -f1,2 | sed “s/^0*//” > lineage.txt
- cut -f2- lineage.txt | awk ‘{if(substr(
1=“”; sub(/^[[:space:]]+/, “”)} print}’ > ranknames.txt - paste lineage.txt ranknames.txt >conjunt_ranknames.txt
9.All the output *.txt files are available in Figshare folder for your check. Customize the file seqtab.nochim_pooling.rds.
The R script script_1_CP.R reads the rds object seqtab.nochim_pooling.rds (obtained in Basic Protocol 1). This is the abundance table (see Table 1) where you can find the DNA sequence for each ASV retrieved. Then the ASV sequences are enumerated with letter-number coding and written to the file ASV_code_sequence.txt.
10.Create the ASVID_DNAseq.rds file from the ASV_code_sequence.txt.
- Run the Rscript asvid_dna.seq.R.
11.Transform the tabulate of the ASV_code_sequence.txt to a new line.
- cat ASV_code_sequence.txt | tr “\t” “\n” > ASV_code_sequence.fasta
12.Linearize the multifasta file.
- awk ‘{if(NR==1) {print
0 ∼ /^>/) {print “\n” 0}}}’ ASV_code_sequence.fasta > ASV_code_sequence_linear.fasta
13.Split the multifasta file (ASV_code_sequence_linear.fasta)
Run the bash script split_fasta.sh in the directory that contains the multifasta file ASV_code_sequence_linear.fasta.
You can do this through the following:
bash splitfasta.sh ASV_code_sequence_linear.fasta, or directly run the commands in the terminal. As you can see, we use the multifasta file as an argument of the bash script.
14.Move all the independent fasta files to a new directory:
- mkdir fasta_files
- mv *.fasta./fasta_files
15.Run BLASTN for all ASV sequences using the custom database obtained in step 7.
- In the terminal, run: bash blastn.sh
This is a long process, so we also provide an alternative script to run parallel tasks on an SGE cluster: blastn-sge.sh
Find the output in the path defined in the previous blastn.sh file.
Check that the number of blasted.txt files coincide with the number of input fasta files. In our case the number is 4617.For example, in the folder that contains the output file, run the next command:
- ls *blasted.txt | wc -l
16.Concatenate all blasted.txt files in a single file.
- cat *blasted.txt >> blastresults.txt
17.Select columns 1, 2 and 4 from the BLASTn output:
- cat blastresults.txt | cut -f1,2,4 > blastotab.txt
Column 1 states for ASV code, column 2 for the sequence identification number and column 4 is the identical percentage of blastn output towards the query sequence (ASV sequence).
Column 2 still appears as 0 in the front of the NCBI taxonomy code, which must be removed. Remove and reorder this to create the final output from BLAST:
- cat blastotab.txt | cut -f1 > column1.txt
- cat blastotab.txt | cut -f2 | sed “s/^0*//” > column2.txt
- cat blastotab.txt | cut -f3 > column3.txt
- paste column1.txt column2.txt column3.txt > blasttotab_def.txt
18.Perform the taxonomical assignment.
- Run Rscript blast_assignment.R
Through the abovementioned R script, an automatic lineage selection per ASV is proposed based on having a pident value >99.5% and taking into account if the lineage defined for BLAST has the maximal resolution (species level).
Output_generated: blastresults_CP.txt
From the initial 4617 ASV sequences, 3811 were blasted to some lineage of the custom BLASTN database. After the blast_assignment.R depuration, 3096 ASV complies with the filters applied to the R script to obtain more confident taxonomical assignments.
19.Manual check and deduplication.
The output of blast for some ASV may not be unique, and more than one species share the same DNA sequence. Deduplication of these cases can only be performed manually because there are many possible situations, and it requires some knowledge on bacterial taxonomy (e.g., Latin knowledge and habitat-specific microbiota lineages).
From the blastresults_CP.txt create a copy where a manual check is done (let's name it, e.g., blastresults_CP_selected.txt). Consider that we do not have to check all the 3096 ASV manually included in blastresults_CP.txt; we only must check the ASV with more than one lineage classification (due to BLAST identical pident value).
For ease of use, open blastresults_CP_selected.txt in office software (such as LibreOffice Calc or Microsoft Excel). By using the conditional formatting-based tool on duplicated ASV numbers you can highlight the ASV to accurate the lineage manually (Figure 2).
After the use of the conditional formatting-based tool, remove the column deep.

Basic Protocol 3: ASV TAXONOMICAL ASSIGNMENT USING NCBI RefSeq TARGETED LOCI PROJECT DATABASE
This protocol uses the public 16S database from NCBI to classify the ASV sequences obtained from the DADA2 pipeline. For this purpose, we use the k-mer taxonomical classification algorithm of Kraken2 and Bracken2, creating a custom database.
Necessary Resources
Hardware
- Linux, MacOS, or Windows (with Subsystem for Linux) operating system, with sufficient available random-access memory (RAM) and disk space (see Strategic Planning)
Software
- The following software must be installed and available in the PATH environment variable to be executable as a binary system:
- Bracken2 (v2.2): (https://github.com/jenniferlu717/Bracken)
- Kraken2 (v2.0.8-beta): (https://github.com/DerrickWood/kraken2)
- Python (v3.9.12): (https://www.python.org/downloads/)
- R (v4.1.2): (https://cran.r-project.org/bin/windows/base/old/4.1.2/)
- Rstudio (v1.4.1106): (https://posit.co/download/rstudio-desktop/)
- We encourage reading the Kraken2 and Bracken 2 user manuals.
- R packages:
- dplyr (v1.1.1): (https://cran.r-project.org/web/packages/dplyr/index.html)
- readxl (v1.4.2): (https://cran.r-project.org/web/packages/readxl/index.html)
Files and sample files
- Available on Figshare
1.Download NCBI RefSeq Targeted Loci project databases.
- wget https://ftp.ncbi.nlm.nih.gov/refseq/TargetedLoci/Archaea/archaea.16SrRNA.fna.gz
- wget https://ftp.ncbi.nlm.nih.gov/refseq/TargetedLoci/Bacteria/bacteria.16SrRNA.fna.gz
For traceability in your study, annotate the download date because NCBI updates the database weekly.
Decompress both downloaded files to obtain fna extension:
- gunzip -d archaea.16SrRNA.fna.gz
- gunzip -d bacteria.16SrRNA.fna.gz
2.Create the custom Kraken2 database.
- mkdir customdatabase
3.Create the Kraken2 custom database only with the two fna files downloaded above from the NCBI RefSeq Targeted Loci.
- kraken2-build --download-taxonomy --db /PATH/customdatabase
This command requires an internet connection to allow kraken2 to download the taxonomy file. Once this first step is finished, execute the next commands to build the custom database with the downloaded fna files:
- for f in *.fna; do kraken2-build --add-to-library $f --db /PATH/customdatabase; done
NOTE: Please verify that the directory where you execute the command ‘for f in *.fna’ contains only the two *.fna files indicated on step 1 and no additional files with the .fna extension.
Once finished, a new library subdirectory will have been created inside custom database directory.
To complete the creation of the Kraken2 custom database, use the following command. You can also execute this command directly in the terminal:kraken2-build --build --threads 28 --db customdatabase
Three files with extension *.k2d will be created (which are essential to work the Kraken2 algorithm downstream).
4.Create Bracken2 k-mers for the custom Kraken2 database.
- bracken-build -d customdatabase -t 28 -k 35 -l 150
For 200 bp kmer:
- bracken-build -d customdatabase -t 28 -k 35 -l 200
For 240 bp kmer:
- bracken-build -d customdatabase -t 28 -k 35 -l 240
Once finished, check that inside the customdatabase directory, you have the next following files: database150mers.kraken, database150mers.kmer_distrib, database200mers.kraken, database200mers.kmer_distrib, database240mers.kraken and , database240mers.kmer_distrib.
5.Run Kraken2.
6.Perform Bracken analysis for the 150 bp kmer.
- bash bracken_refseq.sh
We want to transform the Kraken + Bracken output format to a more friendly (metaphlan format) output. We use the following bash scripts, two Python scripts downloaded from the Krakentools webpage (https://github.com/jenniferlu717/KrakenTools). Indeed, both Python scripts are available in Figshare.
7.Transform each Bracken report to the Metaphlan format.
- bash krakentools_1_refseq.sh
Merge all metaphlan format files into one for downstream analysis. The Python script combine_mpa.py is used within the following bash script.
- bash krakentools_2_refseq.sh
A txt file is generated (combined_allranks_mpa_refseq.txt).
8.Perform last changes/details on the output generated in order to obtain the R file ASV_lineage_refseq.rds with some convenient reformatting of the lineage established with Kraken 2 and Bracken 2 for each ASV using NCBI RefSeq Targeted Loci as a taxonomical database. To achieve this:
- Run Rscript script_refseqafterdada2.R
Basic Protocol 4: DEFINITIVE SELECTION OF LINEAGE AMONG THE THREE METHODS
From the previous protocols, we obtained the lineage for each ASV on the DADA2 pipeline (Basic Protocol 1) from the custom BLASTN database (Basic Protocol 2) and 16S NCBI public databases (Basic Protocol 3). This protocol combines the outputs to consider all our lineage information in three ways: analyze them, compare them, and find common patterns. The aim of this protocol is to obtain a more reliable lineage for each ASV after the output comparison of the three methods. Moreover, this protocol presents an extra final step to ensure the update and homogenization of taxonomic lineage ranks according to NCBI taxonomy (ftp.ncbi.nlm.nih.gov/pub/taxonomy/) using myTAI and taxonomizr R packages. In addition, it shows how to construct the phyloseq object, which is the bridge between bioinformatics and statistical analysis in microbiome data.
Necessary Resources
Hardware
- Linux, MacOS, or Windows (with Subsystem for Linux) operating system, with sufficient available random-access memory (RAM) and disk space (see Strategic Planning)
Software
- The following software must be installed and available in your PATH environment variable to be executable as a system binary:
- R (v4.1.2): (https://cran.r-project.org/bin/windows/base/old/4.1.2/)
- Rstudio (v1.4.1106): (https://posit.co/download/rstudio-desktop/)
- R packages:
- data.table (v1.14.8): (https://cran.r-project.org/web/packages/data.table/index.html)
- dplyr (v1.1.1): (https://cran.r-project.org/web/packages/dplyr/index.html)
- myTAI (v0.9.3): (https://cran.r-project.org/web/packages/myTAI/index.html)
- openxlsx (v4.2.5.2): (https://cran.r-project.org/web/packages/openxlsx/index.html)
- plyr (v1.8.8): (https://cran.r-project.org/web/packages/plyr/index.html)
- stringr (v1.5.0): (https://cran.r-project.org/web/packages/stringr/index.html)
- taxize (v0.9.100): (https://cran.r-project.org/web/packages/taxize/index.html)
- tidyr (v1.3.0): (https://cran.r-project.org/web/packages/tidyr/index.html)
- taxonomizr (v0.10.2): (https://cran.r-project.org/web/packages/taxonomizr/index.html)
Files
- ASV_lineage_refseq.rds, blastresults_CP_selected.txt, dada2lineage_ASVDNA.rds and ASVID_DNAseq.rds (all of them available on Figshare)
Sample files
- See Figshare
1.Select the lineage among the three methods.
- Run Rscript script_pre_phyloseq_16S_CP.R
2.Perform last lineage check and create the phyloseq object.
GUIDELINES FOR UNDERSTANDING RESULTS
For all the script runs, the standard output and standard error are merged, and it is crucial to check for potential error messages. If error messages exist, the results are unreliable, and the error(s) must be resolved before running the next downstream step.
The results retrieved after running BLASTN must be treated through R code blast_assignment.R (as detailed in Basic Protocol 2) to retain all information. There is no manipulation in the output txt files from BLASTN running, only inspection. To remind the meaning of each column of the BLASTN results in the txt files, re-open the bash script blastn.sh where you can find the column names: qseqid (query seq-id); sseqid (subject seq-id); stitle (subject title); pident (percentage of identical matches); qcovs (query coverage per subject); length (alignment length); mismatch (number of mismatches); gapopen (number of gap openings); qstart (start of alignment in query); qend (end of alignment in query); sstart (start of alignment in subject); send (end of alignment in subject); qframe (query frame); sframe (subject frame); frames (query and subject frames separated by a “/”); evalue (expect value); bitscore (bit score); qseq (aligned part of query sequence); and, sseq (aligned part of subject sequence).
The blastresults_CP.txt file obtained from the Basic Protocol 2 must be checked manually. Use the recommendations stated in that protocol, and, in case of doubt, check if the species determined is an inhabitant of the environment of your study.
Protocol 3, the Kraken2 results are saved into two folders (outputs and reports). The txt files in outputs folders are not used for Bracken2 and downstream analysis. Python commands from KrakenTools are used to obtain the Kraken2 result in a Metaphlan format which is easier to manipulate in R, at least for us.
COMMENTARY
Background Information
Due to the nature of 16S rRNA sequencing as NGS technology (based on short reads) and the identical/highly homologous between some species (Gwak & Rho, 2020; Hiergeist et al., 2023), most microbiome studies use the genus rank level as the most resolution taxonomic level. On some occasions, to overcome this limitation and try to classify up to species rank, the tool BLASTN was used (Bazinet et al., 2018; Boisseau et al., 2023) with different parameter configurations due to the absence of a standardized protocol to classify species in 16S rRNA experiments. The most common strategy is to use some taxonomical reference databases integrated into the microbiome bioinformatic pipelines, such as Qiime2 and DADA2. However, a small percentage of ASV is classified at the species level.
Therefore, looking for a strategy to increase the rate of species level classified without compromising the misidentifications of their prediction is interesting to obtain more information from the same analysis that allows finding out specific species of a particular genus implied in some phenotype studied, or at least, to narrow to some species from a genus if a non-unique specie could be established.
First, the standard bioinformatic DADA2 pipeline is described in Basic Protocol 1 and is considered the pattern (the mother method). Basic Protocol 2 illustrates the DADA2 ASV taxonomical classification based on a custom BLASTN database built from the SILVA reference database (also used in Basic Protocol 1) using a confident and robust E-value of 1e-50. Finally, Basic Protocol 3 describes the taxonomical classification of DADA2—ASV using the 16S reference database from NCBI RefSeq Targeted Loci and Kraken2 and Bracken2 as classifier algorithms. Through an automatic process (only a few manual checks are needed), the definitive taxonomic lineage for each ASV has been decided from the reference method (DADA2) and the other two methods (Blast and NCBI RefSeq).
The primary limitation of the current workflow lies in the fact that the 16S databases used for taxonomical profiling are not specifically tailored to the human gut ecosystem. However, this limitation is not inherently problematic, as the protocol is adaptable for various disciplines within microbiota research, including soil microbiology, animal studies, and human research, among others.
Nonetheless, researchers should exercise caution in cases where BLASTN results in different taxonomic lineages with the same percentage identity value, as well as discrepancies among the three methods presented in the manuscript. In such situations, researchers should draw upon their prior experience in their respective research fields and seek relevant literature to confirm whether the uncertain species has been documented within the specific habitat under investigation. Apart from this 3-method approach, two methods have been studied and implemented to improve the 16Sr RNA species classification. One way attempted to classify the DADA2 taxonomically—ASV towards the Unified Human Gastrointestinal Genome (UHGG) v2.0 database (Almeida et al., 2021) using the Kraken2 and Bracken2 as a classifier algorithm (as Basic Protocol 3). Nevertheless, this method was abandoned due to the misclassification of ASV DADA2 sequences to non-16S rRNA sequences. UHGG database contains complete genomes and is designed for shotgun metagenomics analysis. Another method assayed but with unsatisfactory results was to create a custom Kraken2 database with the last version of SILVA. By default, Kraken2 incorporates the SILVA database only up to the genus level. For this reason, through a Python code, we force the incorporation of SILVA v138.1 species multifasta in the Kraken2 algorithm. However, no substantial improvement in species classification was achieved compared to the three methods detailed in the present protocol.
The protocol was performed in a 16S rRNA gene sequencing dataset from 156 human gut samples. In the pattern (mother) method (the DADA2 protocol), only 252 out of the initial 4617 ASV could be classified as species, representing 5.5% (Fig. 4). In contrast, the proposed method based on the output of three methods (DADA2, custom BLASTN and NCBI RefSeq Targeted Loci) classified 1754 ASV as species from 4344 curated ASV in total, increasing the percentage of species classified to 40.4% or 38% if referred to the original 4617 ASV. Analyzing the source of the new species assignments by method, BLAST provided 1371 ASV classified up to species, while 1001 species were retrieved from NCBI RefSeq method.
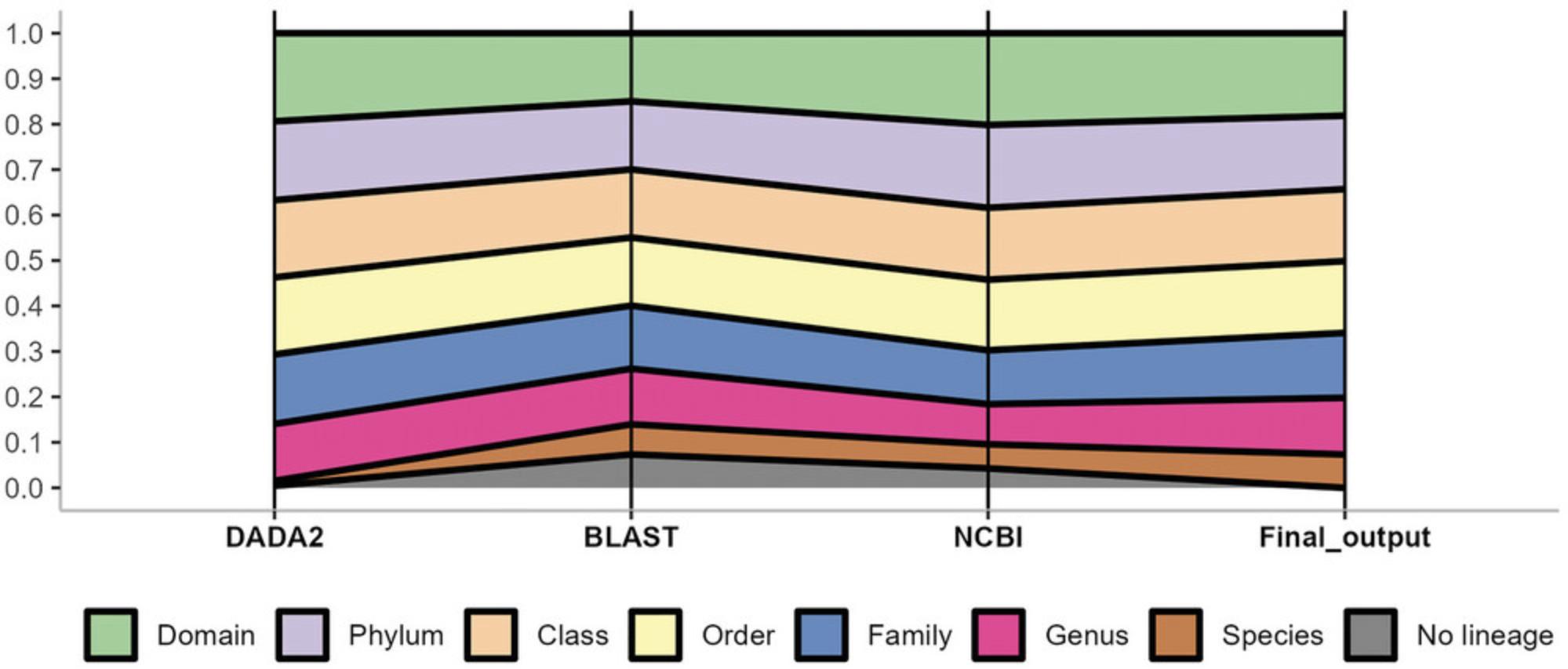
Furthermore, as can be seen in Figure 4, for the most resolution taxonomical ranks (e.g., Genus and Species level) the proposed strategy presented in this manuscript (use of three protocols instead of only one gold standard such as DADA2) allowed to obtain more lineages classified to those levels. In relation to the higher ranks, DADA2 (the pattern method) classified more ASV in comparison to the other two methods (BLAST and NCBI RefSeq) but the corresponding final output values are not so far from DADA2. Finally, DADA2 presented the lowest number of ASV that could not be classified either to Bacteria and/or Archaea (103). Nevertheless, the final output (considering the three methods) presented 267 ASV not classified to Archaea/Bacteria or misclassified to the non-human gut, Mitochondria, and Chloroplast lineages, which is markedly lower than the percentage presented by BLAST and NCBI methodology (1515 and 808 ASV, respectively).
Therefore, and in conclusion, the present protocol increased the practical eight times ASV classifications at the species level by incorporating the information retrieved from two additional methods (BLAST and NCBI RefSeq) apart from the pattern method of DADA2. Then, this methodology could be of interest to research groups that use the 16S rRNA gene sequencing technology in their metagenomic studies.
Critical Parameters
Software's version
Different versions of DADA2, BLAST, Kraken2, and Bracken2 could change the output of the results despite using the same parameters detailed in the present protocol. Therefore, it is crucial to annotate the version of each software used.
Reference databases and lineage nomenclature
The use of other reference databases will completely change the ASV assignment. Furthermore, due to the frequent updates of Bacteria and Archaea in NCBI RefSeq, it is essential to save the download date to reproduce the results. In our current protocol, we have utilized R packages, specifically myTAI and taxonomizr, to update the ancient lineage names from the SILVA v138.1 database to align with the latest NCBI taxonomy database. (https://ncbiinsights.ncbi.nlm.nih.gov/2022/11/14/prokaryotic-phylum-name-changes/). This update ensures that our taxonomy information incorporates the most recent revisions in nomenclature.
Troubleshooting
Table 2 summarizes the common errors documented during the processes detailed in the protocols.
| Problem | Possible cause | Solution |
|---|---|---|
| BLAST Database creation error: Error: Duplicate seq_ids are found | Some headers of the fasta files or multifasta files used to construct a custom BLAST database are identical. | Check that all the headers of the sequences that you want to include in the custom BLAST database are unique. You can use the check_no_duplicates.py to detect it. |
| Can't find taxonomy/subdirectory in the database directory, exiting | Check the bash script running the Kraken2 that the pathway to database is not included in whole detail. | Check that you have included the entire Kraken2 database pathway (from the root). |
| Error: Bad Request (HTTP 400) | Internet failure communication with NCBI (using myTAI package) | Restart the process or subset the process into smaller tasks. |
| Retrieval of taxonomical lineages from species that have more than one possibility (e.g., Escherichia coli/Shigella sonnei, myTAI only considers Shigella sonnei) | Some bug of myTAI R package, unknown origin. | When using the myTAI R package (as stated on the R script) check if the row names (ancient lineage names contain some symbol such as “/” or “-“) and compare to the rank level that is updating if it works well. Check the R script to view an example. |
Time Considerations
Running all of the protocols on the 16S rRNA amplicon data from 156 human fecal samples data showcased herein will take approximately 30 hr 36 min to complete: 24 hr 36 min for Basic Protocol 1, 1 hr 30 min-2 hr for Basic Protocol 2, 3 hr for Basic Protocol 3, and 40-60 min for Basic Protocol 4. The bulk of the time for Basic Protocol 1 (12 hr) is spent in the phylogenetic tree building followed by the denoising step (10 hr), while most of the time for Basic Protocol 2 is spent on the manual check of blast assignment (1 hr-1hr 30 min). For Basic Protocol 3, the running time could be lower in function to the internet connection to The National Center for Biotechnology (NCBI) server that the R package myTAI uses. An analogous scenario occurs for Basic Protocol 4 and the steps required for taxonomizr R package installation account for the main run-time in the last protocol.
The exact time required for each protocol will vary based on the specifications of the computer used to execute the commands. For steps where multiple threads are supported, increasing the number of CPU threads will generally reduce the run-time.
Acknowledgments
D.B-C. received a post-doctoral fellowship from Instituto de Salud Carlos III—grant CD21/00094 (co-funded by European Social Fund. ESF investing in your future). AG-S was supported by the grant FI19/00221 from Instituto de Salud Carlos III (co-funded by European Social Fund. ESF investing in your future) and by the Horizon 2020 grant agreement No 874662 to the HEAP consortium from the European Union. B.R.-S received a pre-doctoral grant from Instituto de Salud Carlos III -grant PFIS FI21/00056 (co-founded by European Union).
VM received grants from Instituto de Salud Carlos III I, co-funded by FEDER funds –a way to build Europe– grants PI17-00092 and PI20/01439. We thank the Spanish Association Against Cancer (AECC) Scientific Foundation.
Author Contributions
David Bars-Cortina : Conceptualization; data curation; formal analysis; investigation; methodology; software; validation; writing—original draft; writing—review and editing. Ferran Moratalla-Navarro : Formal analysis; methodology; software; supervision; writing—review and editing. Ainhoa Garcia-Serrano : Formal analysis; methodology; writing—review and editing. Núria Mach : Methodology; writing—review and editing. Lois Riobó-Mayo : Methodology; writing—review and editing. Jordi Vea-Barbany : Software; writing—review and editing. Blanca Rius-Sansalvador : Methodology; writing—review and editing. Silvia Murcia : Writing—review and editing. Mireia Obón-Santacana : Writing—review and editing. Victor Moreno : Conceptualization; funding acquisition; supervision; writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
All the data used in the protocols are available on Figshare: https://doi.org/10.6084/m9.figshare.23471240.v1
Literature Cited
- Almeida, A., Nayfach, S., Boland, M., Strozzi, F., Beracochea, M., Shi, Z. J., Pollard, K. S., Sakharova, E., Parks, D. H., Hugenholtz, P., Segata, N., Kyrpides, N. C., & Finn, R. D. (2021). A unified catalog of 204,938 reference genomes from the human gut microbiome. Nature Biotechnology , 39, 105–114. https://doi.org/10.1038/s41587-020-0603-3
- Bazinet, A. L., Ondov, B. D., Sommer, D. D., & Ratnayake, S. (2018). BLAST-based validation of metagenomic sequence assignments. PeerJ , 6, e4892. https://doi.org/10.7717/peerj.4892
- Boisseau, M., Dhorne-Pollet, S., Bars-Cortina, D., Courtot, É., Serreau, D., Annonay, G., Lluch, J., Gesbert, A., Reigner, F., Sallé, G., & Mach, N. (2023). Species interactions, stability, and resilience of the gut microbiota - Helminth assemblage in horses. iScience , 26, 106044. https://doi.org/10.1016/j.isci.2023.106044
- Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., & Caporaso, J. G. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology , 37, 852–857. https://doi.org/10.1038/s41587-019-0209-9
- Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., & Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods , 13, 581–583. https://doi.org/10.1038/nmeth.3869
- Chakravorty, S., Helb, D., Burday, M., Connell, N., & Alland, D. (2007). A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of Microbiological Methods , 69, 330–339. https://doi.org/10.1016/j.mimet.2007.02.005
- Gwak, H.-J., & Rho, M. (2020). Data-driven modeling for species-level taxonomic assignment from 16S rRNA: Application to human microbiomes. Frontiers in Microbiology , 11, 570825. https://doi.org/10.3389/fmicb.2020.570825
- Hiergeist, A., Ruelle, J., Emler, S., & Gessner, A. (2023). Reliability of species detection in 16S microbiome analysis: Comparison of five widely used pipelines and recommendations for a more standardized approach. PloS One , 18, e0280870. https://doi.org/10.1371/journal.pone.0280870
- López-Aladid, R., Fernández-Barat, L., Alcaraz-Serrano, V., Bueno-Freire, L., Vázquez, N., Pastor-Ibáñez, R., Palomeque, A., Oscanoa, P., & Torres, A. (2023). Determining the most accurate 16S rRNA hypervariable region for taxonomic identification from respiratory samples. Scientific Reports , 13, 3974. https://doi.org/10.1038/s41598-023-30764-z
- Lu, J., Breitwieser, F. P., Thielen, P., & Salzberg, S. L. (2017). Bracken: Estimating species abundance in metagenomics data. PeerJ Computer Science , 3, e104. https://doi.org/10.7717/peerj-cs.104
- McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., Andersen, G. L., Knight, R., & Hugenholtz, P. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME Journal , 6, 610–618. https://doi.org/10.1038/ismej.2011.139
- Navas-Molina, J. A., Peralta-Sánchez, J. M., González, A., McMurdie, P. J., Vázquez-Baeza, Y., Xu, Z., Ursell, L. K., Lauber, C., Zhou, H., Song, S. J., Huntley, J., Ackermann, G. L., Berg-Lyons, D., Holmes, S., Caporaso, J. G., & Knight, R. (2013). Advancing our understanding of the human microbiome using QIIME. Methods in Enzymology , 531, 371–444. https://doi.org/10.1016/B978-0-12-407863-5.00019-8
- Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., & Glöckner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research , 41, D590–D596. https://doi.org/10.1093/nar/gks1219
- Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., van Horn, D. J., & Weber, C. F. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology , 75, 7537–7541. https://doi.org/10.1128/AEM.01541-09
- Wang, Q., Garrity, G. M., Tiedje, J. M., & Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology , 73, 5261–5267. https://doi.org/10.1128/AEM.00062-07
- Wood, D. E., Lu, J., & Langmead, B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biology , 20, 257. https://doi.org/10.1186/s13059-019-1891-0
Citing Literature
Number of times cited according to CrossRef: 1
- Elies Ramon, Mireia Obón-Santacana, Olfat Khannous-Lleiffe, Ester Saus, Toni Gabaldón, Elisabet Guinó, David Bars-Cortina, Gemma Ibáñez-Sanz, Lorena Rodríguez-Alonso, Alfredo Mata, Ana García-Rodríguez, Victor Moreno, Performance of a Shotgun Prediction Model for Colorectal Cancer When Using 16S rRNA Sequencing Data, International Journal of Molecular Sciences, 10.3390/ijms25021181, 25 , 2, (1181), (2024).

