High-throughput cultivation and identification of bacteria from the plant root microbiota
Jingying Zhang, Yong-Xin Liu, Xiaoxuan Guo, Yuan Qin, Ruben Garrido-Oter, Paul Schulze-Lefert, Yang Bai
Published: 2021-01-12 DOI: 10.1038/s41596-020-00444-7


















Supplementary information
Reporting Summary
Supplementary Table 1
Primer sequences used for bacterial identification. 16S rRNA gene primers 799F and 1193R are in gray. Well and plate barcodes are in red and blue, respectively. Illumina adapters (P5 and Read1, P7, Index and Read2) are in gray.
Supplementary Table 2
Mapping file.
Supplementary Table 3
Taxonomy and sequences of ASVs.
Supplementary Table 4
Read counts of ASVs in each well (ASV table).
Supplementary Table 5
Five best candidate wells corresponding to each ASV.
Supplementary Table 6
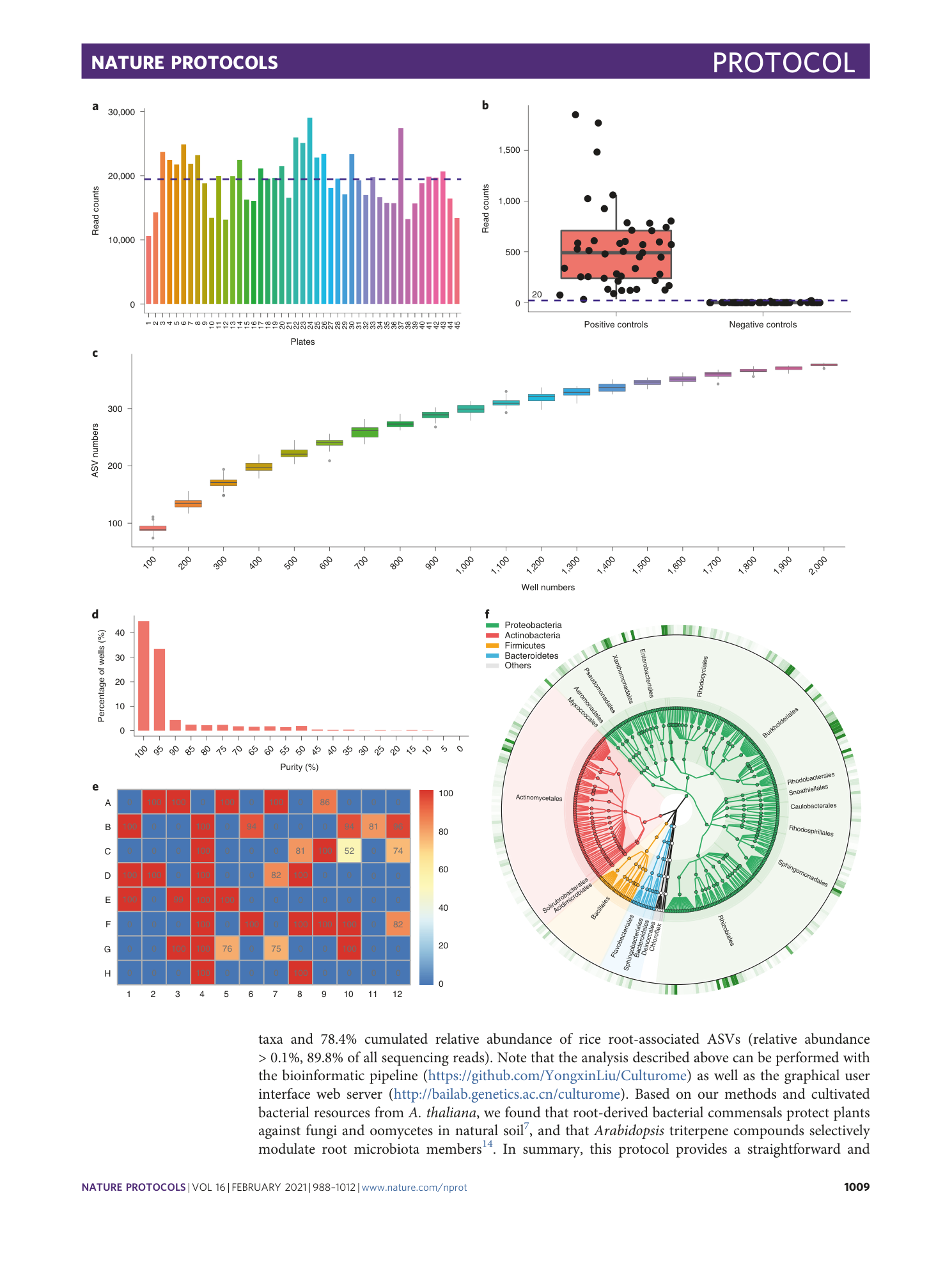
Data to generate anticipated results in Fig. 6.

