cDNA library preparation from total RNA extracts of Single-cell marine protists (e.g. Acantharia, Strombidium basimorphum, and Prymnesium parvum) for transcriptome sequencing
Joost S Mansour, Konstantinos Anestis, Fabrice Not, Uwe John
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Many marine protists are not culturable and therefore challenging to study, nonetheless, they are essential in all marine ecosystems. The development of single-cell techniques is allowing for more marine protists to be studied. Such genomic approaches aim to help to disentangle heterotrophic processes such as phagotrophy from osmotrophy and phototrophic-induced anabolic activities. This information will then support cellular and metabolic modeling by better elucidating the physiological mechanisms and quantifying their importance in different scenarios.However, single-cell protocols and low input RNA kits for transcriptomics are usually made for and tested with mammalian cells, as such the feasibility and efficiency of single-cell transcriptomics on highly diverse mixotrophic protists is not always known. Often single-cell transcriptomics of microbial eukaryotes shows low transcript recovery rates and large variability.
We report on transcriptomic methods that we have successfully performed on single cells of Acantharia, Strombidium basimorphum, and Prymnesium parvum.
This protocol follows up after total RNA extraction (from the protocol at dx.doi.org/10.17504/protocols.io.bp6xmrfn) to prepare cDNA libraries for Illumina sequencing. The described protocol uses the SMART-Seq4 kit (Takara #634891) for cDNA synthesis and amplification, but this can also be successfully performed with the NEBNext kit (NEB #E6421). The NEBNext kit protocol is very similar to the protocol described here and generally the manufacture's protocol can be followed but see the notes at step 4 and step 18 of this protocol, and do the final elution after cDNA purification in 10 mM Tris (pH 8.0).
The subsequent cDNA library is prepared following the

Before start
Total RNA needs to have been extracted (Protocol: dx.doi.org/10.17504/protocols.io.bp6xmrfn) and when possible quantified and quality checked by Bioanalyzer. If Bioanalyzer analysis was possible, only continue with good quality RNA extracts.
- Thaw reagents (except enzymes).
- Allow reagents that need to be at room temperature to incubate at
Room temperature(i.e.and GC nucleic acids purification beads. - Set thermocycler programs and pre-heat thermocyclers.
- For the cDNA purification step Prepare fresh 80% ethanol from
with
Steps
cDNA synthesis preparations
Label for each sample a tube
Prepare a 72°C incubator (e.g. a thermocycler)
Thaw other reagents On ice – except SmartScribe Reverse Transcriptase, take that from the freezer only once needed.
Thaw your RNA samples On ice (as prepared in dx.doi.org/10.17504/protocols.io.bp6xmrfn)
Prepare 10X Reaction Buffer ( RB ), On ice as follows (1 µL is used per sample (adjust as needed, & write down exact volumes):
19µL 1µL
- Mix/vortex and spin down (avoid bubbles)
cDNA synthesis
Take into clean (labeled) 1µL of RNA sample & 1µL of RB
(total 10.5 µL volume, adjust with
Place samples On ice and add 1µL of
add 1µL
Mix gently (vortex) & spin down
Incubate samples at 72°C for 0h 3m 0s
While samples are incubating prepare Master Mix (MM) as below for each sample (+10%; write down exact volumes) On ice
4µL
1µL(pink cap) 0.5µL(white cap)
Immediately after the 3 min 72°C incubation from step 8 put samples On ice for 0h 2m 0s
During this incubation time on ice perform steps 11 and 12.
Preheat thermocycler to 42°C
Take the
2µL
Mix MM by gentle vortex and spin down
Add 7.5µL of the MM to the samples (total volume now 20 µL)
Mix by pipetting and follow with short spindown
Incubate samples in pre-heated Thermocyler with heated lid and the following program:
42°C 1h 30m 0s,
70°C 0h 10m 0s;
4°C
STOPPING POINT - 4°C overnight
cDNA Amplification
Thaw all the reagents (see step 18) On ice except the enzyme
(Vortex and spin down reagents except for enzyme)
Preheat thermocycler to 95°C
Prepare Mastermix (+10%), one sample is as below:
-
25µL -
1µL(green cap) -
3µL -
1µL(take out last minute and mix without vortexing, spin down) -
Mix Master Mix well and gently (finger flick) and spin down
Add 30µL of Mastermix to each sample from cDNA synthesis.Mix well (pipetting) and spin down gently.
Run samples on pre-heated thermocycler with the program:
| A | B | C |
|---|---|---|
| 95°C | 1 min | |
| 98°C | 10 sec | repeat step 2, 18 times |
| 65°C | 30 sec | |
| 68°C | 3 min | |
| 72°C | 10 min | |
| 4°C | forever |
STOPPING POINT 4°C overnight
cDNA cleanup/bead purification
Preparations:
- Label for each sample two tubes
. One tube is used for the cDNA after purification, and one is for an aliquot of the purified cDNA for Bioanalyzer. - Vortex the bead stock well (
), this needs to be very well and evenly mixed - Aliquot beads,
22.5µLx samples (plus extra) - Bring the bead aliquot to
4Room temperaturefor at least0h 30m 0s - Vortex the bead aliquot until evenly mixed
- Prepare fresh 80% EtOH, 400 µL x samples
Add 22.5µL of beads to each sample (amplified cDNA from the previous section)
Mix by pipetting up and down at least 10 times, and vortex
Incubate at 4Room temperature``0h 8m 0s to let cDNA bind to the beads
Pipet and discard the supernatant (72.5 µL), keeping the samples in the magnetic device
Keeping the samples in the magnetic device, add fresh 200µLfresh
Wait 0h 0m 30s
Pipet and discard supernatant containing contaminants (use 100 µL)
Repeat the EtOH washing step for a total of 2 washing steps
Briefly spin the samples to collect liquid off the sides
Place samples back in the magnetic device for 0h 0m 30s, beads will again be collected on the side
Remove all remaining ethanol/supernatant with a pipet (use 10 µL pipet)
Place samples at for minutes. 4Room temperaturefor 0h 2m 0sminutes. (it might take a bit longer)
Until the pellet is no longer shiny, but before a crack appears. It needs to be ‘just’ dry, matte with no shine.
Once the beads are dry add of Elution buffer to all samples 15µL of Elution buffer to all samples to cover the bead pellet
Remove samples from the magnetic device
Mix to re-suspend the beads by (multi)pipetting (can scrap of beads from the side)
Incubate at for (longer) 4Room temperaturefor0h 2m 0s (longer) to rehydrate
Briefly spin the samples to collect liquid off the sides
Place the samples back in the magnetic device for 0h 1m 0s, until the solution is completely clear
Transfer the clear supernatant containing purified cDNA to
Make immediately an aliquot for Bioanalyzer analysis to prevent unnecessary freeze-thawing cycles.
STOPPING POINT - Label and store at -20°C
cDNA Sample verification
Quantify and calculate the concentration of cDNA. This is needed for the next cDNA library procedure.
cDNA library preparation and indexing – Nextera XT
Proceed with cDNA library preparation only for good quality samples from the previous step.
Normalize cDNA samples to 30pg/ul
Dilute each sample of amplified and purified cDNA to 30 pg/µL in either Elution buffer or as per the final step of the used protocol for cDNA purification. Work with a minimum of 1 µL amplified cDNA and a total volume of 5 µL.
Prepare to work very timely for this protocol
- Preheat a PCR thermocycler to
55°C, with preheat lid at 100 °C - Prepare from the
the ATM and NT reagents in sufficient quantity (i.e. 5 ul per sample for each) separated over multiple tubes to facilitate multi-pipetting
Follow the
Refer to pages 7-9 of the Nextera XT manual (https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-05.pdf).
Changes to manufacturer' s protocol:
- Start the tagmentation with
5µLof 30 pg/µl amplified cDNA sample (from step 37) - all steps indicated as "centrifuge at 280 x g at 20 °C for 1 minute" can be substituted short spindown in a tabletop mini-centrifuge.
Store samples at 4°C for up to 2 days or proceed immediately with purification
cDNA library purification
Preparations:
- Vortex the bead stock well (
), this needs to be very well and evenly mixed - Aliquot beads,
30µLx samples (plus extra) - Bring the bead aliquot to
4Room temperaturefor at least0h 30m 0s - Vortex the bead aliquot until evenly mixed
- Prepare fresh 80% EtOH, 400 µL x #samples
Spin down your indexed cDNA samples (total 50 µL)
Add 30 µL of
- Mix by pipetting up and down
- Shake/vortex for
0h 2m 0s
Incubate at 4Room temperature``0h 5m 0s to let cDNA bind to the beads
Briefly spin down and place the samples on a for 0h 5m 0s or longer. Until the liquid appears completely clear and there are no beads in the supernatant.
Pipet and discard the supernatant (80 µL), keeping the samples in the magnetic device
Keeping the samples in the magnetic device, add fresh 200µLfresh
Wait 0h 0m 30s
Pipet and discard supernatant containing contaminants (use 100 µL pipet)
Repeat the EtOH washing step for a total of 2 washing steps
Briefly spin the samples to collect liquid off the sides
Place samples back in the magnetic device for 0h 0m 30s, beads will again be collected on the side
Remove all remaining ethanol/supernatant with a pipet (use 10 µL pipet)
Place samples at for minutes. 4Room temperaturefor 0h 5m 0sminutes.
Until the pellet is no longer shiny, but before a crack appears. It needs to be ‘just’ dry, matte with no shine.
Once the beads are dry add of 52.5µL of
Remove samples from the magnetic device
Mix to re-suspend the beads by (multi)pipetting (can scrap of beads from the side)
Vortex for 0h 2m 0s followed by a very short spindown
Incubate at for 4Room temperaturefor0h 2m 0s to rehydrate
Briefly spin the samples to collect liquid off the sides
Place the samples back in the magnetic device for 0h 2m 0s, until the solution is completely clear
Transfer the clear supernatant (50 uL) containing your purified cDNA library to
Make immediately an aliquot for Bioanalyser analysis to prevent unnecessary freeze-thawing cycles.
STOPPING POINT - Label and store at -20°C for sequencing
cDNA library verification
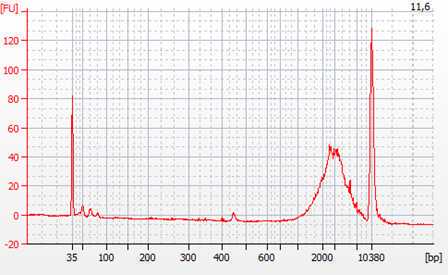
Check the quality of the cDNA libraries by

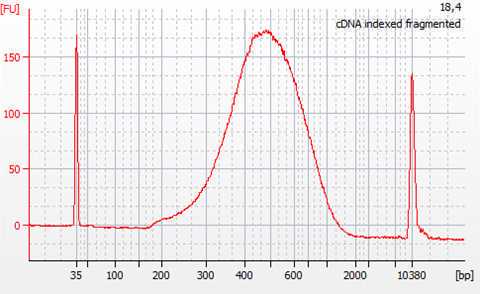
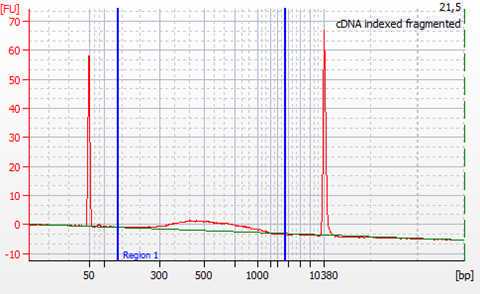
Quantify and calculate the concentration of cDNA by smear analysis. This is needed for the normalization of samples for sequencing.
4.4.3 Follow up steps: library quality control; sample normalization/dilution and pooling for sequencing
The quality and quantity control of the generated cDNA libraries is performed using the Agilent High Sensitivity DNA kit (Agilent #5067-4626). In case primer-dimers or adapters are still present, an additional step of cleaning with magnetic beads is to be performed. A bead to sample ratio of 0.7:1 was found to be efficient in eliminating both primer dimers and remaining adapters.
The cDNA libraries are normalized to equal molarity, as well as fragment size before the final pooling and subsequent sequencing. Calculate nM cDNA of each sample as: nM DNA = [ng/µL] x 106/ (660 x fragment length bp). Where the concentration in ng/µL and the average fragment length in base pairs are obtained from Bioanalyzer smear analysis.
The molarity upon which the cDNA libraries are normalized is determined based on the yield of cDNA, as well as the requirements for the subsequent sequencing (e.g. >0.5 nM). The final pool of all the samples should again be checked using the Bioanalyzer in order to verify that the normalization process was successful.
The pools are ready for Illumina sequencing.