Whole-Genome Amplification of Respiratory Syncytial Virus (RSV) using Illumina CovidSeq reagents for Next-Generation Sequencing
Carlos Davina-Nunez, Sonia Perez-Castro, Montse Godoy-Diz, Benito Regueiro-Garcia
Disclaimer
Abstract
This protocol has been tested for amplification of RSV-positive nasopharyngeal swabs of CT value up to 24 using Seegene Allplex Respiratory Panel (Seegene Inc, Seoul, South Korea). This protocol does not require prior subtyping as it covers RSV-A and RSV-B in the same reaction. Panel of primers is an optimisation of a previously published panel by Wang et al.
This panel was modified to optimise the multiplex PCR, so the whole genome can be amplified in just two PCR reactions. In addition to this, primers have been modified to account for commonly-occurring mutations in the 22-23 season that affect primer-binding areas and were causing suboptimal amplification.
These primers were used to cover the complete hRSV genome (both A and B) by splitting into two pools of non-consecutive amplicons (odd-numbered amplicon primers in one pool, even-numbered amplicon primers in other). This allowed for Whole-genome amplification in two reactions.
Illumina CovidSeq (Illumina Inc, San Diego, USA) reagents were used for the RT-PCR, with a mix previously published for amplification of Influenza RNA and a thermocycling program optimised in our lab. The library preparation part of the protocol was performed according to the Illumina CovidSeq protocol.
Before start
This protocol uses as input RNA extracted from nasopharyngeal swabs after confirmation of RSV infection via RT-PCR. Samples were extracted using the QIASymphony DSP Virus/Pathogen Midi Kit (Qiagen, Hilden, Germany).
Steps
Primer pools preparation
Prepare both primer mixes according to Table 1.
For a final concentration of 10uM: add 1017 ul of Nuclease-Free water to Pool 1 and 1035 ul to Pool 2.
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| A1f | 5 | Wang | ACGSGAAAAAATGCGTACAAC | 1 | 1 |
| A1r | 5 | Wang | GAAGATTGTGCTATACCAAAATGAACA | 1779 | 1 |
| AB3f | 10 | Goya | GCYATGGCAAGACTYAGGAATG | 2897 | 1 |
| A3r | 5 | Wang | GTTTGCYGAGGCTATGAATATGAT | 4826 | 1 |
| A5f | 5 | Wang | GAACAACAGACTACTAGAGATTACCAG | 6374 | 1 |
| A5r | 10 | This publication | AGGAGTTTRCTCATRGCAA | 7929 | 1 |
| A7f | 5 | Wang | AGCTTAGGCTTAAGATGYGGA | 9423 | 1 |
| A7r | 5 | Wang | TGAGTTTGACCTTCCATGAGT | 10997 | 1 |
| A9f | 7 | Wang | GGGTTGGTTCATCTACACAAGAG | 12316 | 1 |
| A9r | 7 | This publication | CGCAATAATAAATTCCCTGCTCC | 14094 | 1 |
| B1f | 5 | Wang | ACGCGAAAAAATGCGTACTACA | 1 | 1 |
| B1r | 5 | Wang | CATTGTTTGCCCTCCTAATTACTG | 1661 | 1 |
| B3r | 5 | Wang | ATAGGGCCAAAATTTGCTTGTG | 4309 | 1 |
| B5f | 5 | Wang | AGTGCAATCTTCCTAACTCTTGC | 5700 | 1 |
| B5r | 5 | Wang | TGATTCCACTTAGTTGGTCTTTGC | 7375 | 1 |
| B7f | 5 | Wang | GGTGAACTGAAATTAGAAGAACCAAC | 8760 | 1 |
| B7r | 5 | Wang | CACCATATCTTGTCAAACTCTCAGG | 10507 | 1 |
| B9f | 7 | Wang | GAACCAACTTACCCTCATGGATT | 11860 | 1 |
| B9r | 7 | Wang | TTCTGGGGTTGGGTGATATAG | 13650 | 1 |
| A2f | 5 | Wang | ACAGGCATGACTCTCCTGAT | 1556 | 2 |
| A2r | 5 | Wang | TTGGGTGTGGATATTTGTTTCAC | 3400 | 2 |
| A4f | 5 | Wang | ACCTGGGACACTCTCAATCA | 4697 | 2 |
| A4r | 5 | Wang | GACATGATAGAGTAACTTTGCTGTCT | 6540 | 2 |
| A6f | 5 | Wang | GTCACGAAGGAATCCTTGCA | 7642 | 2 |
| A6r | 5 | Wang | CCCTCTACCTCTTTTATTATGTAGAACC | 9521 | 2 |
| A8f | 5 | Wang | GGTGTACAATCTCTATTTTCCTGGT | 10704 | 2 |
| A8r | 5 | Wang | CGATTAATAGGGCTAGTATCAAAGTG | 12615 | 2 |
| A10f | 10 | This publication | CRTCTACAATGATTAGAACCAATTAC | 13742 | 2 |
| A10r | 10 | Wang | ACGAGAAAAAAAGTGTCAAAAACTAA | 15225 | 2 |
| B2f | 5 | Wang | CAGRTTAGGAAGGGAAGACACTA | 1316 | 2 |
| B2r | 5 | Wang | CAAGTCACTCAATTTTTTGGAGGTTGG | 2982 | 2 |
| B4f | 10 | Wang | TGGAAGCAYACAGCTACACG | 3943 | 2 |
| B4r | 10 | Wang | CTACATGTYGATTGGTAAAACTCC | 5788 | 2 |
| B6f | 5 | Wang | CCTCTAGTGTTTCCTTCTGATGAG | 7113 | 2 |
| B6r | 5 | Wang | GTTGTAGCAATTTGTTCAGACGAG | 8834 | 2 |
| B8f | 5 | Wang | AAGTTCTCTGAAAGCGACAGATC | 10231 | 2 |
| B8r | 5 | This publication | TAATACTWGGTGATGTTACTCCTAC | 12190 | 2 |
| B10f | 5 | Wang | TAGTCAATCAAGACACAAGTTTGC | 13289 | 2 |
| B10r | 5 | Wang | ACGAGAAAAAAAGTGTCAAAAACTAATG | 15222 | 2 |
Table 1: mix of primers used for amplification. Two mixes are required, one for pool 1 and another for pool 2. References for base number: hRSV/A/England/397/2017 and hRSV/B/Australia/VIC-RCH056/2019 for RSV-A and RSV-B respectively. Citation to the original papers for the primers can be found below.
RT-PCR
Two Master Mixes must be prepared per sample: one for Pool1 and one for Pool 2 (Table 2). Manipulate reagents according to the Illumina CovidSeq Reference Guide.
| A | B | C |
|---|---|---|
| IPM | 15 | 15 |
| FSM | 3.2 | 3.2 |
| RVT | 1 | 1 |
| Nuclease-Free Water | 3.6 | 3.6 |
| Primer pool 1 (10uM) | 1.2 | - |
| Primer pool 2 (10uM) | - | 1.2 |
Table 2: Master mixes required for amplification of the RSV genome. Reaction 1 targets odd-numbered amplicons while reaction 2 targets even-numbered amplicons.
In a PCR tube, mix 20 ul of MasterMix with 5 ul of extracted RNA.
Place all tubes (two per sample) in a thermocycler and run the following program (Table 3):
| A | B | C |
|---|---|---|
| 42º | 60 min | |
| 98º | 2 min | |
| 98º | 15 s | 35 cycles |
| 63º | 7 min | |
| 4º | PAUSE |
Table 3: Thermocycler program for RT-PCR. Indicate 25 ul as volume and heat lid at 99ºC.
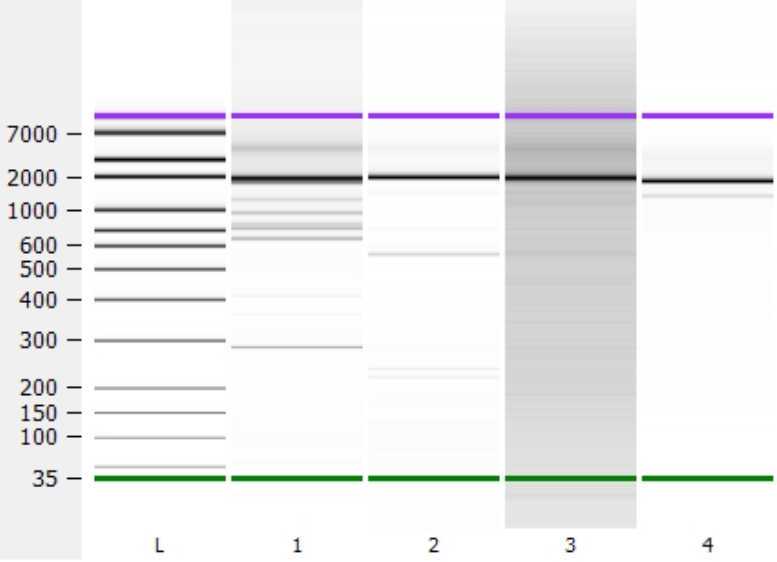
(OPTIONAL) Check RT-PCR result with Agilent Bioanalyzer
Use an Agilent Bioanalyzer to check for amplification peaks. Expect PCR peaks around ~2000 bps.
Library preparation
Mix 10ul of tube one and tube two on each sample for a final 20 ul of PCR product. Follow instructions of the Illumina CovidSeq Reference Guide to generate sequencing-ready libraries.
Recommended: To ensure optimal normalisation, perform the library Clean-up on each tube and normalise individually instead of pooling. This improves normalisation especially in the presence of low-concentration PCR products.
Quantify samples after Clean-up using Qubit Flex and normalise samples.
(OPTIONAL): Check library preparation on an Agilent Bioanalyzer. The pattern expected is the usual post-tagmentation pattern from Illumina libraries with the highest peak around ~330bps.
Expected results
Coverage obtained after an iSeq run with 16 samples: 8 from RSV-A, 7 from RSV-B and a negative control.
Average reads per sample (excluding negative control): 361k
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。https://lh4.googleusercontent.com/FjwTBcX_2JHv13sPkhkt3o2DifOTneaG7w2xTRCkyQewtJDXo1AktipKlNhA7l5HBCJDfWaBxc_-gEVI2UduryGuiPAhbn0I6Y4JFLoTB06tIKDM4M5-pZ5xh_IymoKgnoE0yXfgFIX9b7gSc4aAHEMReferences
The illumina CovidSeq protocol can be found in:
Illumina CovidSeq Reference Guide
The primers found in Table 1 were obtained from:
The Master Mix used for RT-PCR with Illumina CovidSeq was first published in: