Visualization and Enumeration of Ostreococcus via Kohler Transmitted Light/Dark-Field on the Axio A1 Imager Microscope and AxioCam HrC
Lynn Doran
Abstract
Basic setup and usage instructions for visualization of Ostreococcus species on the Axio A1 Imager Microscope using the AxioCam HrC and AxioVision for live and static imaging on the desktop using the Kohler Transmitted Light/Dark-field Method. Captured static images using the AxioCam HrC can then be exported for organism enumeration manually or using ImageJ Cell Counter or equivalent programs.
Before start
Steps
Prepare organisms
Sterilize a laminar flow hood or biological safety cabinet, pipettes, and gloves. Autoclave pipette tips. Sterilize the outside of the culture vial, especially the lid.
Gently rock culture vial to evenly suspend organisms. Using aseptic technique, pipette 10µLof culture into the hemacytometer. Depending on cell density, the culture may need to be diluted with the appropriate culture media before being transferred to hemacytometer.
Setup Microscope Controls
The lever below the condenser light intensity slider controls an iris diaphragm that is used to focus the light beam. Ensure that this lever is pushed towards the back of the microscope and the full light field is visible.

Remove all items from the stage to avoid the possibility of scratching the objective when switching to a higher magnification if the stage is too close for the larger objective. Select the desired objective. Use the wheel to change objectives. Do not use the objective to turn the wheel or you could damage it.


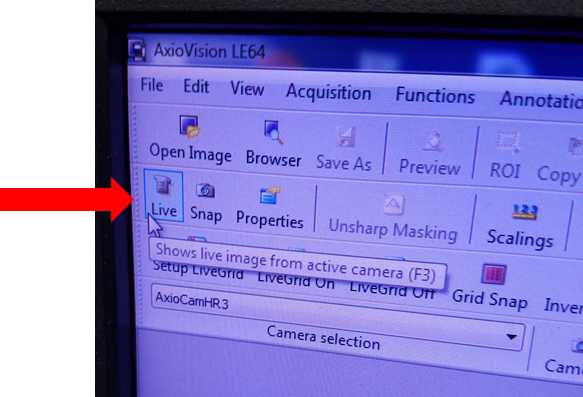
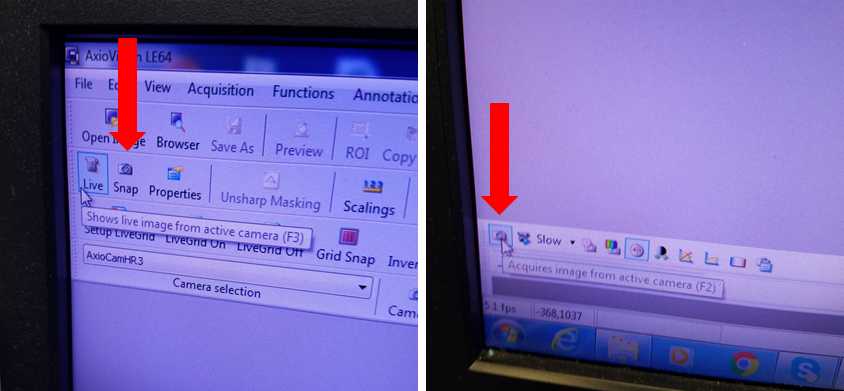
Activate AxioCam HrC Software
Focus Sample
Software image adjustments
If an image appears on the live screen that is not representative of your sample and does not move when you move the platform, an overlay may have been previously saved on the software. To remove the overlay image, right click in the live view and a new window of options will open, select “load factory defaults”.
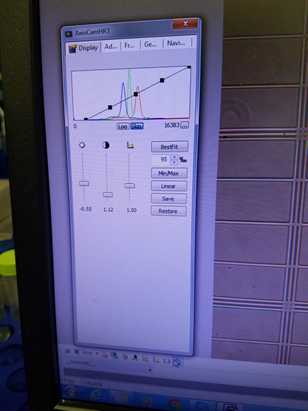
On the second tab, "Adjust", sliding the tone slider to warmer or cooler changes the color hue of the image and may make the organisms easier to identify. Images presented in this protocol have Ostreococcus appearing as a black/green nucleus on an orange hued background with white hemacytometer grid lines. To recreate this color scheme, move the slider to "warmer".
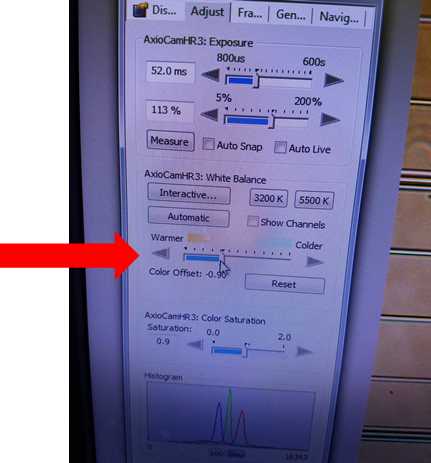
Additional desired image adjustments can be made in the properties menu. For additional image options, please refer to the AxioVision manual provided in the Materials tab.
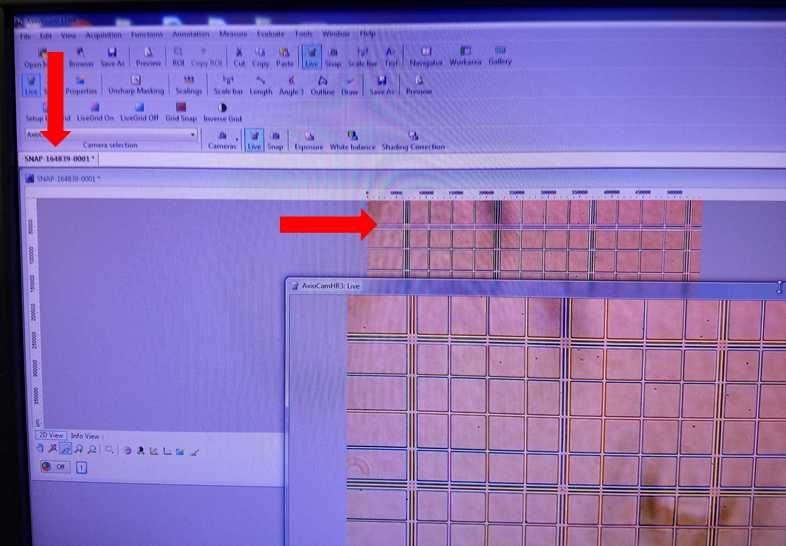
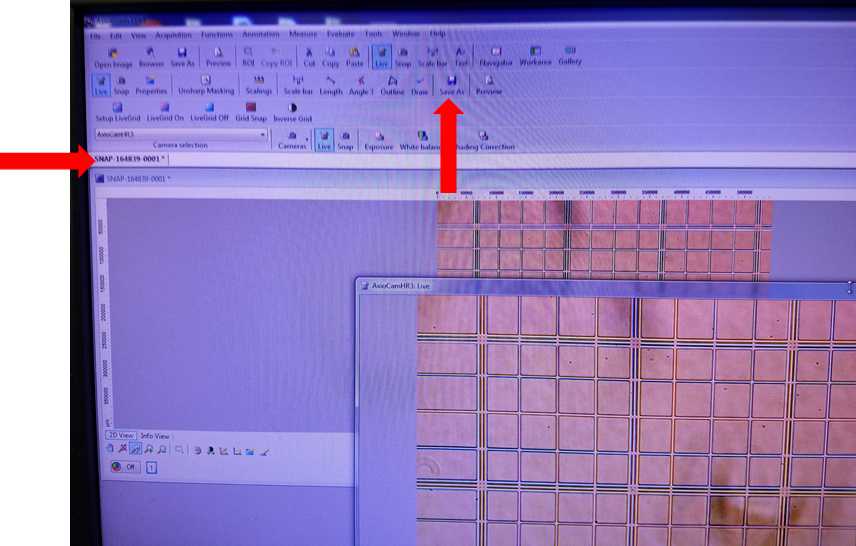
Image Capture
Click on the desired snapshot to perform image analysis in the software such as scaling, angle measurement, or freehand drawing. Image analysis can not be performed on live image. Additional information about post-processing image analysis can be found in the AxioVision manual.
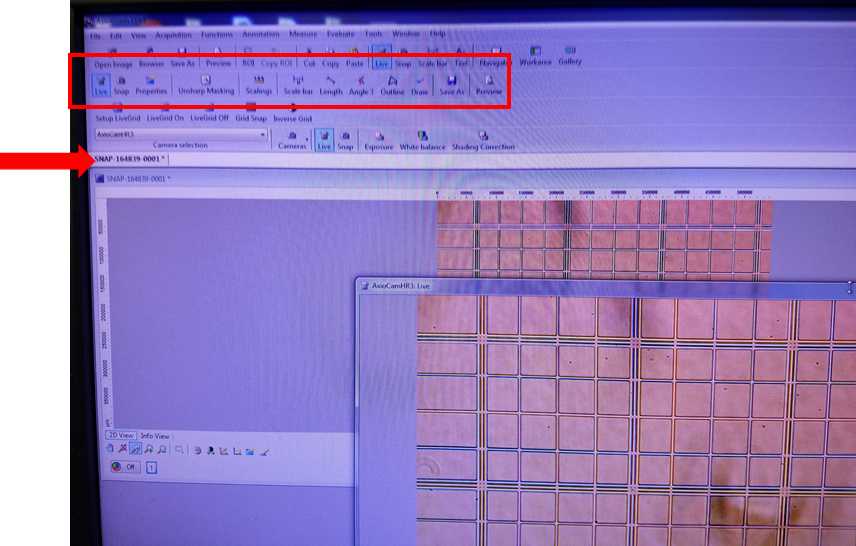
The individual photo files are now able to be transferred to other analysis software such as ImageJ with Cell Counter Add-In for enumeration.