Validation of Genotyping Method for L444P Mice Ear-Clips
David C, Revi Shahar Golan
Abstract
Aim : the genotyping is used to identify if mice are heterozygote (hetero) or Wild-Type (WT), and the aim of the work is to validate the digestion method, and PCR program, the PCR primers, and the interpretation of the results.
General notes : There are two sets of primers – Neo primers to distinguish between WT and hetero samples, and L444P primers to detect L444P protein. Negative and positive samples are used to verify the digestion and PCR, by gel electrophoresis. Sequencing is used to verify the interpretation of the gel electrophoresis.
Steps
Digestion
Prepare Digestion mix: 15µL (Roche, cat 10241100) per 1000µL (Viagen, cat 402-E).
Add 50µL to each ear clip sample.
Heat for 55°C, 5h 0m 0s.
Heat 99°C for 0h 10m 0s.
Cool to 10°C.
Samples can be stored in -20°C until use.
PCR
Prepare PCR master mix (MM) as below ( per sample ):
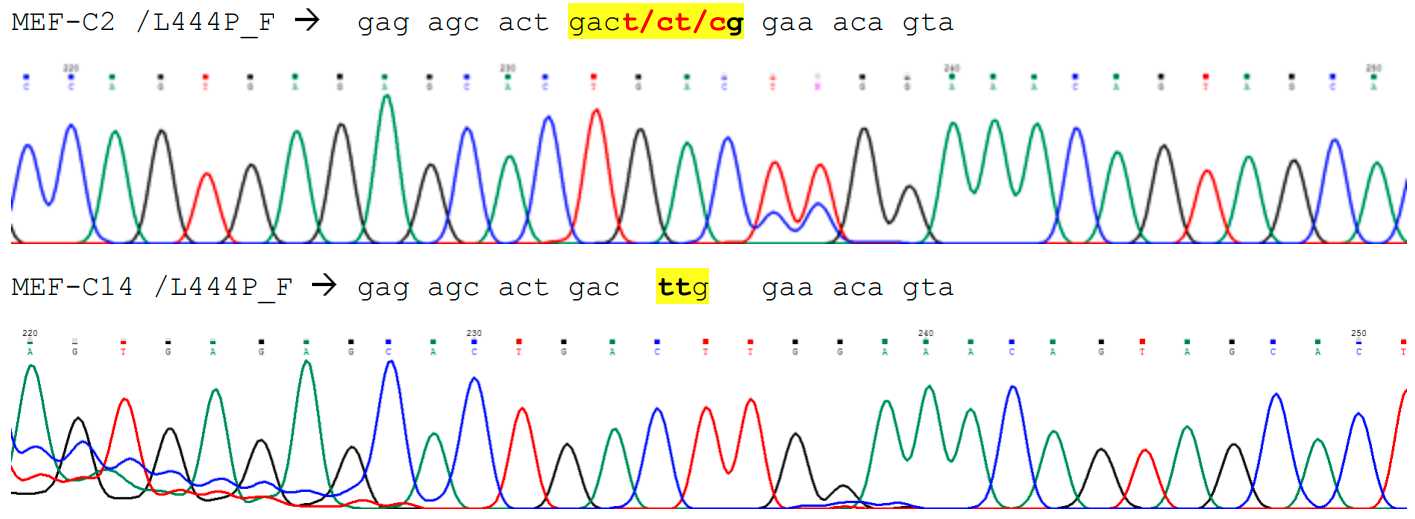
2.5µL(VWR-Peqlab, cat 01-1020)0.25µL(VWR-Peqlab, cat 01-1020)0.5µL(10pmol/ µl)0.5µL(10pmol/ µl)0.5µL(40mM; VWR, cat 5100850-0500)19.25µL(Promega, cat P1193)NotePrimers details: Neo1: 5’GATTGCACGCAGGTTCTCCG3’ Neo2: 5’CCAACGCTATGTCCTGATAG3’ L444P_F: 5’CCCCAGATGACTT GATGCTGG3’ (marked in bold , an extra T, that does not appear on the sequence) L444P_R: 5’CCAGGTCAGGATCTCTGATGG3’
For each sample, add 23.5µL and 1.5µL.
PCR program:
| A | B | C |
|---|---|---|
| Temp | Time | Cycles |
| 94°C | 5 min | |
| 94°C | 30 sec | 35 cycles |
| 55°C | 1 min | |
| 72°C | 1 min | |
| 4°C | forever |
PCR product can be kept in 4°C for the short term, or at -20°C.
Gel electrophoresis
Run PCR products on a pre-cast 2% SYBR-safe E-GEL (ThermoFisher, cat G521802), for 0h 30m 0s, along with a 100bp DNA ladder (NEB, N3231), and a negative control (no DNA, only MM).
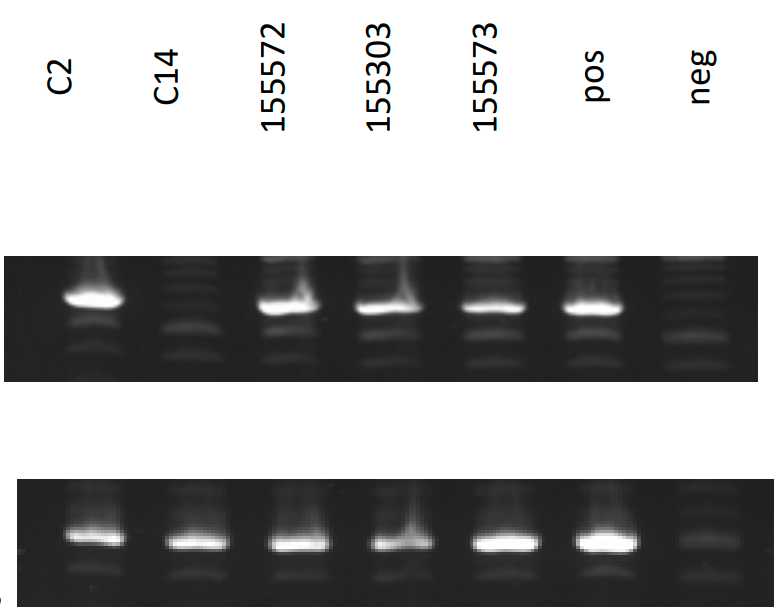
Routine check of ear clips:
Run digested ear clips with NEO primers in the first instance. Then choose one of three options:
- If the ratio wt:hetero is 1:3, there is no need for further tests. Wt animals to be terminated.
- If there are more wt than the 1:3 ratio, this suggested poor DNA for the samples that were not amplified, so need to amplify with L444P primers the samples that did not work, and sequence them.
- If there are much more hetero than the 1:3 ratio, need to sequence all the samples.