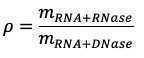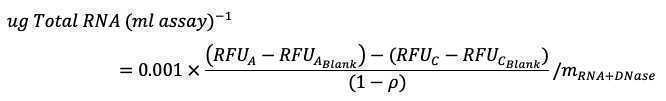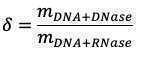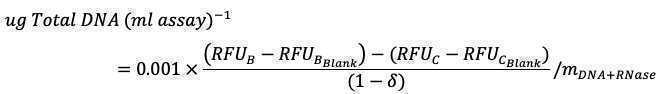Total RNA and DNA from Microalgae (24 samples per day)
Ying-Yu Hu, Zoe V. Finkel
Abstract
Presented herein is a protocol for the extraction and quantification of bulk RNA and DNA from microalgae, adapted from the methodology outlined by Berdalet et al. (2005). RNA and DNA are extracted from microalgae samples and quantified using the fluorochrome SYBR Green II. To ensure accuracy, the concentrations of RNA and DNA standards are determined via absorbance measurements at 260 nm and 320 nm. This additional step aids in maintaining the consistency of the standard curve coefficients (i.e., the slope). The method demonstrates a sensitivity range of approximately 20-700 ng/ml for RNA and 10-700 ng/ml for DNA in the assay.
Before start
Steps
Sample collection
Filter microalgae in liquid media onto polycarbonate filters, using gentle vacuum pressure (130 mmHg).
Rinse filter tunnel with filtered artificial seawater (nutrient free) to avoid sample loss.
Fold the filter with two tweezers:
(1) Fold in half along its diameter, creating a semicircular shape;
(2) Fold once more in the same direction, resulting in a long strip;
(3) Fold once more, halving its length, so that sample is secured.
Place folded filter in 2 mL cryogenic vial.
Blank is not required in this measurement.
Flash-freeze tubes with liquid nitrogen and store at -80°C
Freeze-dry samples. Store at -80°C.
Equipment
| Value | Label |
|---|---|
| FreeZone® 2.5 L Benchtop Freeze Dryers | NAME |
| Labconco® | BRAND |
| 700202000 | SKU |
Primary solutions
Turn on UV light in biosafety cabinet for 0h 15m 0s
Clean working surface with decontamination solution.
Prepare Tris buffer 5mM 8.0
Pour 1M 8.0 Tris into an RNase free 15 mL Falcon tube
Equipment
| Value | Label |
|---|---|
| Falcon® Centrifuge Tubes | NAME |
| Polypropylene, Sterile, 15 mL | TYPE |
| Corning® | BRAND |
| 352096 | SKU |
Directly add 2.5mL 1M``8.0 Tris into 500 mL RNase free water in its original package.
Equipment
| Value | Label |
|---|---|
| BT Barrier Pipet Tips | NAME |
| Pre-Sterile | TYPE |
| Neptune® | BRAND |
| BT1250, BT100, BT10 | SKU |
RNA primary standard solution (200ug/ml )
In the original package, the frozen E. Coli Total RNA is of 1 mg/mL, in which total RNA is 200 ug.
Uncap the original package of E. Coli Total RNA and directly add 800µL Tris buffer (5mM ,8.0) .
Cap the package and invert for a thorough mix.
Aliquot 30 uL by stepper with sterile tip to 600 uL RNase free microtubes. Store at -80°C
Equipment
| Value | Label |
|---|---|
| Finnpipette Stepper Pipette | NAME |
| Thermo Scientific™ | BRAND |
| 4540000 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
Equipment
| Value | Label |
|---|---|
| Finntip stepper pipette tips | NAME |
| 500 ul (sterile) | TYPE |
| Thermo Scientific | BRAND |
| Thermo Scientific™ 9404173 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
DNA primary standard solution (≈ 500ug/ml)
Uncap the original package of Deoxyribonucleic acid from calf thymus and add 2mL Tris buffer (5mM ,8.0).
Cap the package. Do not vortex or sonicate.
Keep the solution in the fridge overnight to completely solubilize the DNA. Gentle reversion is recommended.
Aliquot 10 uL by stepper with sterile tip to 600 uL RNase free microtubes. Store at -20°C
Equipment
| Value | Label |
|---|---|
| Finntip stepper pipette tips | NAME |
| 500 ul (sterile) | TYPE |
| Thermo Scientific | BRAND |
| Thermo Scientific™ 9404173 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
RNase primary stock solution (10mg/ml )
Uncap the original package of Ribonuclease A from bovin pancreas and add 5mL Tris buffer (5mM ,8.0). Cap the package and invert for a thorough mix.
Aliquot 30 uL by stepper with sterile tip to 600 uL RNase free microtubes. Store at -20°C
Equipment
| Value | Label |
|---|---|
| Finntip stepper pipette tips | NAME |
| 500 ul (sterile) | TYPE |
| Thermo Scientific | BRAND |
| Thermo Scientific™ 9404173 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
Equipment
| Value | Label |
|---|---|
| Finntip™ Stepper Pipette Tips | NAME |
| 500 ul (Sterile) | TYPE |
| Thermo Scientific | BRAND |
| 21-377-149 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
DNase primary stock solution (5mg/ml = 10,000 U/mL)
Uncap the original package of Deoxyribonuclease1 and add 1mL Tris buffer (5mM ,8.0) .Cap the package and invert for a thorough mix.
Use reverse pipetting to precisely aliquot 60 uL into 5 mL RNase free tube. Store at -20°C.
Total RNA and DNA extraction
Turn on UV light in biosafety cabinet for 0h 15m 0s
Clean working surface with decontamination solution.
Prepare falcon tubes and tube rack in biosafety cabinet| A | B | | --- | --- | | 5 | 0.5 M EDTA | | 5 | 20% sarcosine | | 50 | 5 mM Tris | | 15 or 50 | 1% STEB |
Equipment
| Value | Label |
|---|---|
| Falcon® Centrifuge Tubes | NAME |
| Polypropylene, Sterile, 15 mL | TYPE |
| Corning® | BRAND |
| 352096 | SKU |
Equipment
| Value | Label |
|---|---|
| Falcon® Centrifuge Tubes | NAME |
| Polypropylene, Sterile, 50 mL | TYPE |
| Corning® | BRAND |
| 352070 | SKU |
Prepare STEB (1% )
Pour sarcosine (20% ) into an RNase free 5 mL falcon tube.
Pour EDTA (0.5M ) into an RNase free 5 mL falcon tube.
Pour Tris buffer (5mM ,8.0) into an RNase free 50 mL falcon tube.
Mix the following ingredients to obtain STEB (1% ):
sarcosine (20% ): 500µL
EDTA (0.5M ): 10µL
Tris buffer (5mM ,8.0): 9mL+490µL
Prepare ice bath
Remove freeze-dried samples from -80ºC freezer and place them On ice.
Add 500µL Tris buffer (5mM ,8.0) and 500µL STEB (1% ) to the bead tube. Place tubes On ice.
Equipment
| Value | Label |
|---|---|
| Lysing tube matrix D | NAME |
| 2 mL | TYPE |
| MP biomedical | BRAND |
| 116913500 | SKU |
| http://www.mpbio.com | LINK |
Rinse forceps by 70% volume ethanol and air dry.
Equipment
| Value | Label |
|---|---|
| Filter forceps | NAME |
| blunt end, stainless steel | TYPE |
| Millipore | BRAND |
| XX6200006P | SKU |
| http://www.emdmillipore.com/ | LINK |
Transfer sample/blank filter into the bead tube by using clean forceps.
Prior to placing sample filter into the bead tube:
Invert the tube then put back On ice.
Turn on refrigerated centrifuge and set the temperature to 4°C.
Equipment
| Value | Label |
|---|---|
| CENTRIFUGE 5430 R | NAME |
| Eppendorf | BRAND |
| MP2231000510 | SKU |
Disrupt samples on the bead mill at 6.5 m/s. Equipment
| Value | Label |
|---|---|
| Fastprep-24 5G™ Sample Preparation Instrument | NAME |
| MP Biomedicals | BRAND |
| 116005500 | SKU |
Keep tubes On ice. Check the label on each tube, restore the label if it fades.
Disrupt samples on the bead mill at 6.5 m/s.
Keep tubes On ice. Check the label on each tube, restore the label if it fades.
Disrupt samples on the bead mill at 6.5 m/s
Keep tubes On ice. Check the label on each tube, restore the label if it fades.
Disrupt samples on the bead mill at 6.5 m/s.
Continuously shake homogenate in a multi-head vortex at the highest speed for 1h 0m 0s Room temperature
Centrifuge extracted samples 10000x g,4°C
In the biosafety cabinet, prepare 3XN 2 mL RNase free microtubes, N = number of samples.
In the biosafety cabinet, transfer all extract to their corresponding microtubes.
Centrifuge extracted samples 10000x g,4°C
In the biosafety cabinet, transfer supernatant as much as possible to their corresponding microtubes, avoid disturbing the debris.
In the biosafety cabinet, transfer supernatant for the assay.
Invert the tube and mix thoroughly, then aspire 100µL of supernatant using the reverse pipetting technique.
Dispense this 100 uL into the 2 mL microtube.
Return any remaining extract in the tip back to the extraction tube.
Place the aliquot and the remaining extract into two separate boxes, i.e., one for aliquot only, the other one for remaining extract.
Store both boxes of samples at -80°C.
Assay (Be prepared: it is a full working day procedure)
Prepare ice bath.
Turn on UV light in biosafety cabinet for 0h 15m 0s
Clean working surface with decontamination solution.
Prepare falcon tubes, microtubes and tube racks in biosafety cabinet| A | B | C | | --- | --- | --- | | 4 | 5 mL falcon tubes | 1 M MgCl2 | | | | 1 M CaCl2 | | | | Working solution B (WS-B) | | | | Working solution C (WS-C) | | 1 | 50 mL falcon tube | 5 mM Tris buffer | | 1 | 15 mL falcon tubes | 0.05% STEB | | 6 | 2 mL RNase free tubes | RNase working solution | | | | RNA tertiary standard solution | | | | DNA tertiary standard solution | | | | 900 mM MgCl2 | | | | 900 mM CaCl2 | | | | Sybr green working solution (SG-II WS) | | 24 | 2 mL RNase free tubes | RNA standard solutions for RNA standard curves | | | | DNA standard solutions for DNA standard curves | | 3XN (N = Number of samples) | 2 mL RNase free tubes | Diluted samples | | 4 | Microtube racks | Tubes of 2 mL in Set 1 | | | | Tubes of 2 mL in Set A | | | | Tubes of 2 mL in Set B | | | | Tubes of 2 mL in Set C | | 1 | Tube racks | Falcon tubes |
Equipment
| Value | Label |
|---|---|
| Screw-Cap Centrifuge Tube | NAME |
| 5 mL | TYPE |
| VWR | BRAND |
| 10002-738 | SKU |
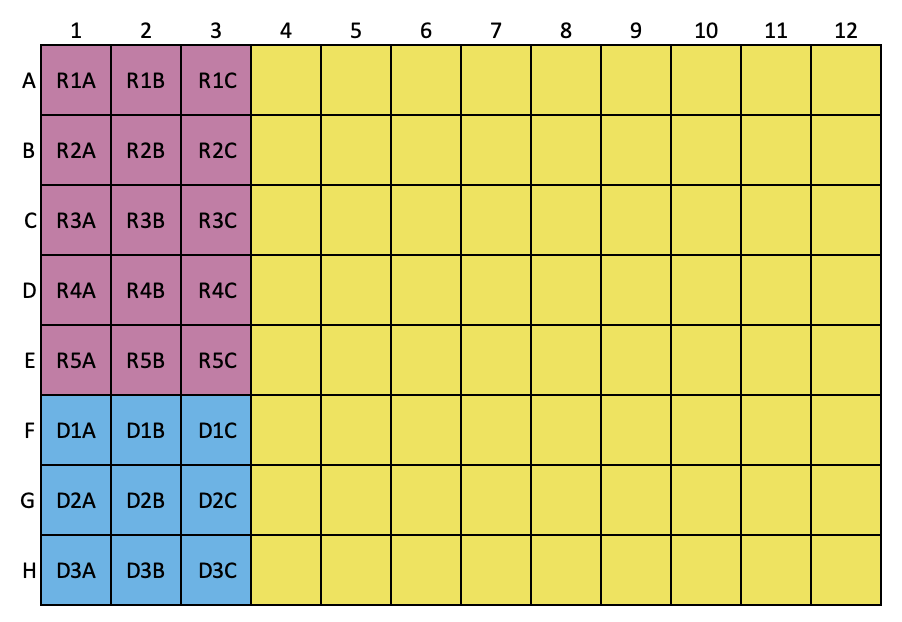
Organize and label the tubes as shown below, log sample numbers into the tube rack layout.
Set 1:
This tube rack holds sample extract (100 uL) to be further diluted.

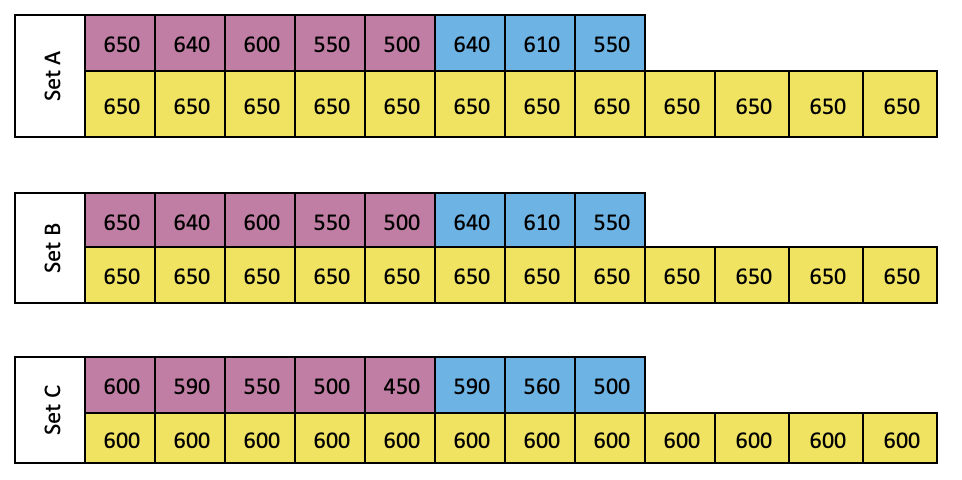
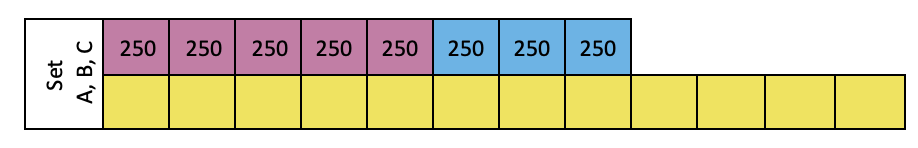
Set A, B and C:
In microtube rack, label 2 mL tubes for RNA (marked in pink), DNA (marked in blue) standard solutions and samples (marked in yellow)
Set A is for working solution A (WS-A) treatment, i.e. treated with DNase
Set B is for working solution B (WS-B) treatment, i.e. treated with RNase
Set C is for working solution A (WS-A) and C (WS-C) treatment, i.e. treated with DNase and RNase
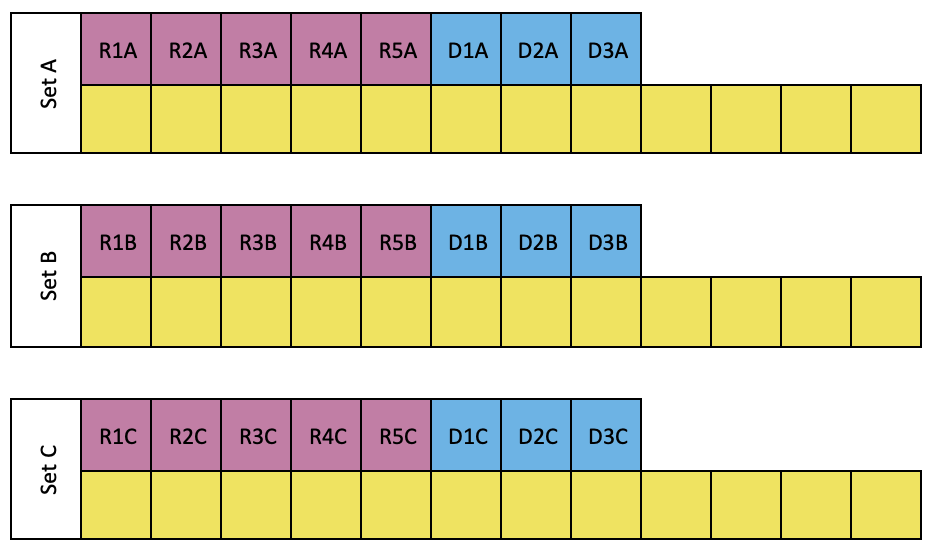
Label tubes for reagents as following.
Follow the sheet, add Tris buffer (5mM ,8.0) to the reagent tubes:
| A | B |
|---|---|
| SG-II WS | 1000+250 |
| WS-B | 2X1000+820 |
| WS-C | 2X1000+940 |
| RNase | 380 |
| 900 mM MgCl2 | 40 |
| 900 mM CaCl2 | 40 |
| RNA tertiary | 690X2 |
| DNA tertiary | 960 |
| 0.05% STEB | 9X1000 + 500 |
Thaw Sybr green II at room temperature
Prepare STEB (0.05% )
Add 500µL STEB (1% ) to 0.05% STEB tube, and invert to mix.
Place sample extract, RNase and DNase primary stock solutions, RNA and DNA primary standard solutions -20On ice.
Important technique for accurately preparing standards and working solutions
Prepare DNA secondary standard solution 25ug/ml
Add 190µL 5 mM Tris buffer into DNA primary tube.
Prepare DNA tertiary standard solution 1ug/ml
Mix DNA secondary standard solution by aspiring up and down several times with pipet.
Transfer 40µL DNA secondary solution (25ug/ml) to DNA tertiary standard tube.
Keep 37On ice.
Prepare RNA secondary standard solution 25ug/ml
Add 210µL Tris buffer into RNA primary tube.
Prepare RNA tertiary standard solution 2ug/ml
Mix RNA secondary standard solution by aspiring up and down several times with pipet.
Transfer 120µL RNA secondary solution (25ug/ml) to RNA tertiary standard tube and mix.
Keep 37On ice.
Remove the DNA and RNA secondary out of the biosafety cabinet.
Before loading the samples, use pipet tip to aspire and dispense multiple times for thorough mix.
Use reverse pipetting:
Load 4 ul DNA and RNA secondary onto the μdrop plate, in duplicate.
Equipment
| Value | Label |
|---|---|
| µDrop™ Plates | NAME |
| Thermo Scientific | BRAND |
| N12391 | SKU |
| https://www.lifetechnologies.com | LINK |
Equipment
| Value | Label |
|---|---|
| Varioskan LUX Multimode Microplate Reader | NAME |
| Thermo Fisher | BRAND |
| VL0L00D0 | SKU |
Read absorbance at 260 and 320 nm.
DNA_primary concentration (μg/ml) = (Abs260-Abs320)x 50 μg/ml x (10mm/0.49 mm) X DF
Where, DF=20.
The DNA primary concentration should be around 500 ug/mL.
If the DNA primary concentration is less than 400 (reading from udrop is less than 0.055) or higher than 600 ug/mL, repeat to . Pay more attention to the solution mixing of primary DNA solution.
RNA_primary concentration (μg/ml) = (Abs260-Abs320)x 40 μg/ml x (10mm/0.49 mm) X DF
Where, DF=8.
The DNA primary concentration should be around 200 ug/mL.
If the DNA primary concentration is less than 150 (reading from udrop is less than 0.055) or higher than 250 ug/mL, repeat to . Pay more attention to the solution mixing of primary RNA solution.
Turn on shaker/incubator and set temperature to 37°C.
Equipment
| Value | Label |
|---|---|
| SHAKING INCUBATOR | NAME |
| 71L | TYPE |
| Corning® LSE™ | BRAND |
| 6753 | SKU |
Prepare 900mM MgCl2
Pour 1M MgCl2solution into 5 mL RNase free Falcon tube
Transfer 360µL 1M MgCl2solution into 900 mM MgCl2tube
Add 60µL 900mM MgCl2 to WS-B
Prepare 900mM CaCl2
Pour 1M CaCl2solution into 5 mL RNase free Falcon tube
Transfer 360µL 1M CaCl2solution into 900 mM CaCl2tube
Add 60µL 900mM CaCl2 to WS-B
Prepare SG-II WS
Centrifuge one tube of SG-II concentrate at Room temperature 13000rpm to deposit DMSO.
Wrap SG-II WS tube with foil, transfer supernatant supernatant of SYBR Green II 10,000X concentrate to SG-II WS tube in biosafety cabinet ( 8.75ul per 1.25 mL Tris)
Check absorbance of SG-II WS:
In a transparent microplate, load
(1) 200 uL Tris buffer as blank
(2) 10 uL SG-II WS and 190 uL Tris buffer
Read absorbance at 480 nm, the value after subtracted by blank shall be no higher than 0.21
Prepare RNase working solution 0.5mg/ml
Add 20µL RNase primary stock solution (10mg/ml) to RNase tube
Thoroughly mix RNase tube and then transfer 60µL 0.5mg/ml RNase working solution to WS-B.
Keep WS-B 37On ice .
Thoroughly mix RNase tube and then transfer 60µL 0.5mg/ml RNase working solution to WS-C.
Keep WS-C 37On ice .
Label DNase tube with "WS-A", add 2X1000+820 mL 5 mM Tris buffer into the tube.
Add 60µL 900mM MgCl2 to WS-A
Add 60µL 900mM CaCl2 to WS-A.
Keep WS-A 37On ice.
Use reverse pipetting: load 50µL WS-A to tubes in Set A .
Use reverse pipetting: load 50µL WS-A to tubes Set C .
Use reverse pipetting: load 50µL WS-B to tubes in Set B .
Use reverse pipetting: load 50µL WS-C to tubes in Set C .
Thoroughly mix sample prior to transferring.
Follow the layout below to add diluted samples, RNA tertiary and DNA tertiary into Set A, B and C. The unit of volume is uL.
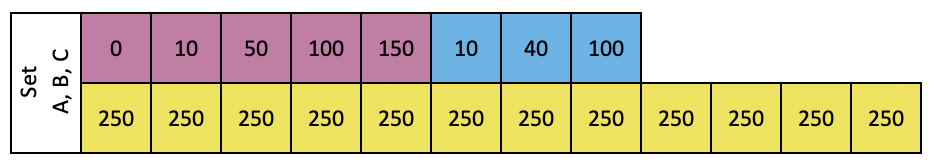
Load diluted samples to each corresponding tubes (marked in yellow) in Set A, B and C .
Add RNA tertiary standard to tubes (marked in pink) in Set A, B and C .
Add DNA tertiary standard to tubes (marked in blue) in Set A, B and C .
Place all tubes into the shaker/incubator at 37°C, continuously shaking at 200 RPM for 0h 20m 0s.
After incubation, Invert each tube for thorough mixing, then place them into the freezer for 2 min to stop the reaction.
Fluorescence measurement
Remove samples out of the freezer and allow to reach Room temperature before loading the microplate.
Adhere black film on the top of a microplate lid.
Equipment
| Value | Label |
|---|---|
| Black Vinyl Films for Fluorescence and Photoprotection | NAME |
| VWR | BRAND |
| 89087-692 | SKU |
Equipment
| Value | Label |
|---|---|
| Microplate Lids | NAME |
| Polystyrene | TYPE |
| Greiner Bio-One | BRAND |
| 07000288 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
Load 10µL SG-II WS to each well by either 0.5 mL stepper or 10 uL pipet. Cover the plate with the black-film lid.
Equipment
| Value | Label |
|---|---|
| 96-Well Black Microplates | NAME |
| Polystyrene | TYPE |
| Greiner Bio-One | BRAND |
| 655076 | SKU |
Shake black film covered microplate at 37Room temperature for 1h 30m 0s
Setup microplate reader:Plate: Greiner F bottom chimney well PP 96 well;Shake: Continuous 5s at 600 rpmFluorescence bandwidth: 12 nm Exciation: 490 nmEmission: 520 nm Equipment
| Value | Label |
|---|---|
| Varioskan LUX Multimode Microplate Reader | NAME |
| Thermo Fisher | BRAND |
| VL0L00D0 | SKU |
Read fluorescence and export data to excel sheet.
In the fume hood, dispose any waste with SG-II into fluorescence stain waste container (some stain waste has DMSO solvent).
Calculation
RNA standard curve
Concentrations of RNA standards in the microplate: Use measured RNA primary concentration instead of 200 ug/mL:
| A | B | C | D |
|---|---|---|---|
| 30 | 210 |
| A | B | C | D |
|---|---|---|---|
| 120 | 1380 |
| A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|
| R1 | 0.00 | 650.00 | 250.00 | 50.00 | 190.00 | 10.00 | 0.00 |
| R2 | 10.00 | 640.00 | 250.00 | 50.00 | 190.00 | 10.00 | ~20.00 |
| R3 | 50.00 | 600.00 | 250.00 | 50.00 | 190.00 | 10.00 | ~100.00 |
| R4 | 100.00 | 550.00 | 250.00 | 50.00 | 190.00 | 10.00 | ~200.00 |
| R5 | 150.00 | 500.00 | 250.00 | 50.00 | 190.00 | 10.00 | ~300.00 |
Slope of fluorescence in Set A vs concentration of RNA standard gives m RNA+DNase (~0.024)
Slope of fluorescence in Set B vs concentration of RNA standard gives mRNA+RNase
DNA standard curve
Concentrations of DNA standards in the microplate: Use measured DNA primary concentration instead of 500 ug/mL:
| A | B | C | D |
|---|---|---|---|
| 10 | 190 |
| A | B | C | D |
|---|---|---|---|
| 40 | 960 |
| A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|
| R1 | 0 | 650 | 250 | 50 | 190 | 10 | 0 |
| D1 | 10 | 640 | 250 | 50 | 190 | 10 | ~10 |
| D2 | 40 | 610 | 250 | 50 | 190 | 10 | ~40 |
| D3 | 100 | 550 | 250 | 50 | 190 | 10 | ~100 |
Slope of fluorescence in Set A vs concentration of DNA standard gives m DNA+DNase
Slope of fluorescence in Set B vs concentration of DNA standard gives m DNA+RNase (~0.13)
Dilution factor=40
If,
- Sample is extracted by 1 mL extraction reagent
- In Set 1, sample is diluted to 100/1000
- In Set 3, diluted by Tris and all working solutions to 250/950
- In microplate, diluted by SG-II WS to 190/200