Three-Dimensional Cell Culture of Epididymal Basal Cells and Organoids: A Novel Tool for Toxicology
Julie Dufresne, Julie Dufresne, Mary Gregory, Mary Gregory, Laurie Pinel, Laurie Pinel, Daniel G. Cyr, Daniel G. Cyr
Abstract
Spermatozoa are formed in the testis but must transit through the epididymis to acquire motility and the ability to fertilize. The epididymis is a single convoluted tubule comprising several anatomically and physiologically distinct regions. The pseudostratified epithelium consists of multiple cell types, including principal cells, clear cells, narrow cells, and apical cells, that line the lumen of the epididymis. Basal cells are present at the base of the epithelium, and halo cells, which includes macrophages/monocytes, mononuclear phagocytes, and T lymphocytes, are also present in the epithelium. Several aspects of this complex spermatozoan maturation process are well established, but a great deal remains poorly understood. Given that dysfunction of the epididymis has been associated with male infertility, in vitro tools to study epididymal function and epididymal sperm maturation are required. Our lab and others have previously developed human, rat, and mouse epithelial principal cell lines, which have been used to address certain questions, such as about the regulation of junctional proteins in the epididymis, as well as the toxicity of nonylphenols. Given that the epididymal epithelium comprises multiple cell types, however, a 3D in vitro model provides a more comprehensive and realistic tool that can be used to study and elucidate the multiple aspects of epididymal function. The purpose of this article is to provide detailed information regarding the preparation, maintenance, passaging, and immunofluorescent staining of rat epididymal organoids derived from adult basal cells, which we have demonstrated to be a type of adult stem cell in the rat epididymis. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Isolation of epididymal cells
Basic Protocol 2 : Magnetic activated cell sorting and isolation of basal cells
Basic Protocol 3 : Preparation and culture of epididymal basal cell organoids
Basic Protocol 4 : Passage of epididymal basal cell organoids
Basic Protocol 5 : Freezing and thawing of epididymal basal cell organoids
Basic Protocol 6 : Immunofluorescent staining of epididymal basal cell organoids
INTRODUCTION
While spermatozoa are formed in the epithelium of the seminiferous tubules of the testis, they lack motility and the ability to fertilize (Bedford et al., 1973; Orgebin-Crist, 1968; Robaire et al., 2015; Turner, 2002 2008). These functions are acquired as spermatozoa transit through the lumen of the epididymis via a process referred to as sperm maturation. Although this process is complex and remains poorly understood, several aspects of the maturation process are well established (Cornwall, 2009; Robaire et al., 2015; Turner, 1995, 2008). These include secretion of proteins by epididymal principal cells that bind to maturing sperm; apocrine secretion of exosomes, known as epididymosomes, which transport cargo to sperm; and variations in pH levels along the epididymis (Barrachina et al., 2022; Breton & Brown, 2013; Breton et al., 1996; Sharma et al., 2016; Sullivan, 2015). The blood-epididymis barrier (BEB), created by tight junctional complexes between cells of the epididymal epithelium, plays a central role in creating the luminal environment that is necessary to promote sperm maturation (Agarwal & Hoffer, 1989; Cyr et al., 2018; Dube & Cyr, 2012; Franca et al., 2012; Mital et al., 2011). It has been shown that dysfunction of the epididymis is associated with infertility (Dube et al., 2008; Dube, Dufresne, et al., 2010; Dube, Hermo, et al., 2010). Gene expression data have shown altered gene expression in epididymides of infertile men with either non-obstructive or obstructive azoospermia (Dube et al., 2008; Dube, Hermo, et al., 2010). Hence, the maintenance of a normal epididymal epithelium is requisite for sperm maturation.
The epididymis is composed of a single convoluted tubule that is divided into four major regions in most species (Jelinsky et al., 2007; Robaire & Hermo, 1988; Robaire et al., 2015). These include the initial segment, the caput, and the corpus and cauda regions (Robaire & Hermo, 1988; Turner et al., 2003). Each of these regions is both anatomically and physiologically distinct. The epithelium is pseudostratified and is composed of multiple cell types (Fig. 1) (Hermo et al., 1992; Sullivan et al., 2019; Sun & Flickinger, 1980). These include principal cells, clear cells, and apical cells that line the lumen of the epididymis, as well as narrow cells that are present in the initial segment only (Adamali & Hermo, 1996; Robaire et al., 2015). Basal cells are present at the base of the epithelium, and halo cells, which include macrophages/monocytes, mononuclear phagocytes, and T lymphocytes, are also present in the epithelium (Fig. 1) (Battistone et al., 2023; Battistone et al., 2020; Breton et al., 2019; Robaire et al., 2015; Shum et al., 2013; Voisin et al., 2020).
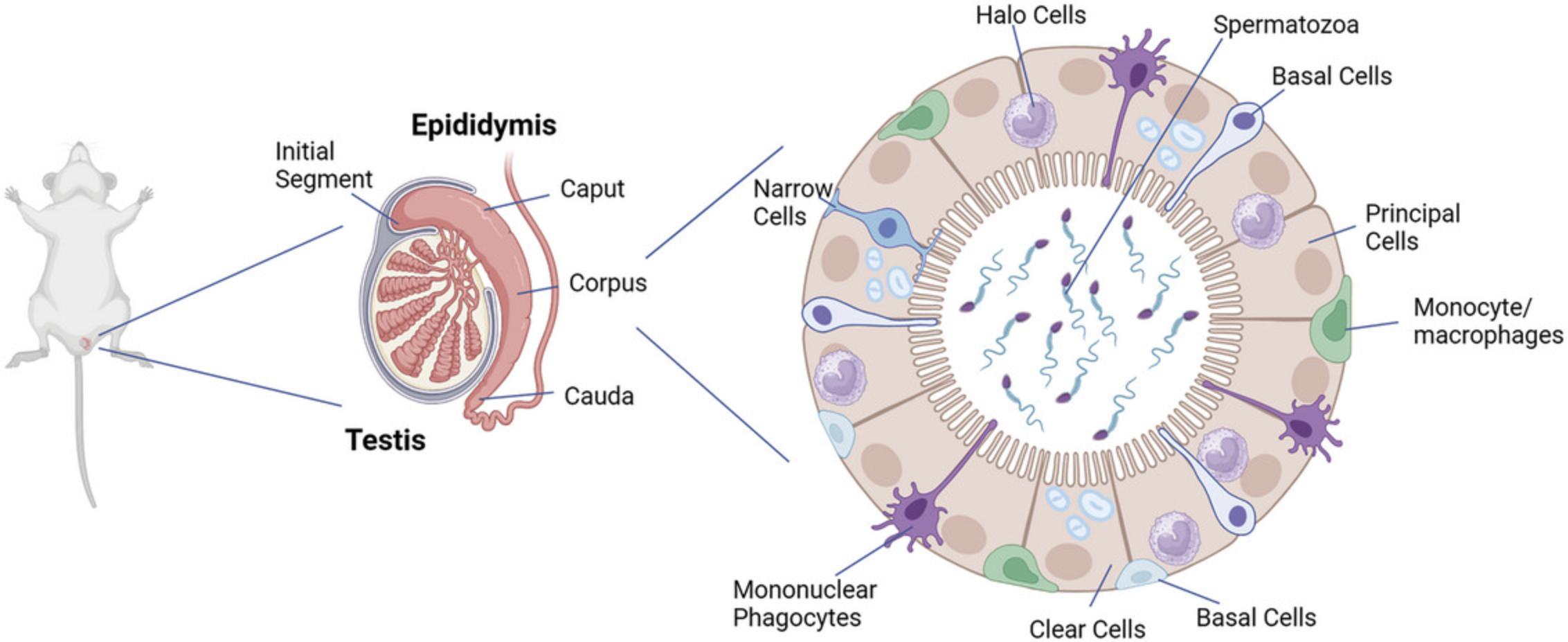
Our lab and others have developed human (Dube, Dufresne, et al., 2010; Dube, Hermo, et al., 2010), rat (Dufresne et al., 2005), and mouse (Araki et al., 2002; Sipila et al., 2004) epithelial principal cell lines, which have been used to address certain questions, such as about the regulation of junctional proteins in the epididymis (Adam & Cyr, 2016; Dufresne & Cyr, 2007; Gregory & Cyr, 2019), as well as the toxicity of nonylphenols (Jones & Cyr, 2011). Given that the epididymal epithelium consists of multiple cell types, however, a 3D in vitro model provides a more comprehensive and realistic tool that can be used to study and elucidate the multiple aspects of epididymal function.
In order to fully understand the role and functions of basal cells within the epididymal epithelium and their contributions to the maturation of spermatozoa, we have developed protocols to isolate basal cells from the rat epididymis (Mandon et al., 2015). Using a variety of cellular and molecular markers, we showed that we could isolate basal cells from the rat epididymis using magnetic bead separation and anti-CD49f antibody. We were able to consistently achieve >90% purity in our preparations (Mandon et al., 2015). We subsequently developed an in vitro 3D organoid culture protocol for these isolated basal cells, which could then be differentiated into other epididymal cell types (Pinel & Cyr, 2021).
The development of basal cell–derived organoids represents a major advance in our understanding of epididymal function and provides a unique model to understand the regulation of epididymal function (Cyr & Pinel, 2022). The following protocols describe the isolation of rat epididymal basal cells (Basic Protocols 1 and 2) and the 3D cell culture for the formation and differentiation of basal cell organoids (Basic Protocol 3), as well as the passaging (Basic Protocol 4), freezing (Basic Protocol 5), and immunofluorescent staining (Basic Protocol 6) of these organoids.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals.
NOTE : All steps should be done under aseptic conditions, such as in a laminar flow hood, unless indicated otherwise.
Basic Protocol 1: ISOLATION OF EPIDIDYMAL CELLS
This protocol produces a cell suspension containing all epididymal cell types. Epididymides from adult rats are harvested, cleaned, cut into fragments, and digested with collagenase type I. The cells obtained after digestion are dissociated with trypsin, and aggregates are removed with cell strainers. At the end of the protocol, we expect to obtain a total of 10 to 25 million cells (per four adult rats) that contain mostly the principal cells of the epididymis but also the basal cells, which can be isolated with Basic Protocol 2.
Materials
-
Sprague-Dawley rats, 48 days old (n = 4; Charles River Laboratories)
-
DMEM/Ham F12 supplemented with penicillin-streptomycin (50 U/ml and 50 µg/ml; MilliporeSigma, cat. no. D6421 and P0781), 37°C
-
Digestion medium: DMEM/Ham F12 supplemented with penicillin-streptomycin (50 U/ml and 50 µg/ml; MilliporeSigma, cat. no. D6421 and P0781) and collagenase Type I (2 mg/ml; Gibco/Thermo Fisher Scientific/Life Technologies, cat. no. 17100017; add collagenase fresh on day of experiment), 37°C
-
1× trypsin-EDTA (Gibco/Thermo Fisher Scientific, cat. no. 15400054)
-
DMEM/Ham F12 supplemented with penicillin-streptomycin (50 U/ml and 50 µg/ml) and 5% (v/v) fetal bovine serum (FBS; Corning, cat. no. 35-077-CV), 37°C
-
Trypan blue solution (Gibco/Thermo Fisher Scientific, cat. no. 15250061)
-
Surgical instruments (blunt-tip fine tweezers, small fine-point scissors, and scalpel), sterile
-
50-ml conical tubes, sterile
-
100-mm Petri dishes, sterile
-
125-ml Erlenmeyer flasks, sterile
-
Aluminum foil
-
Parafilm
-
37°C shaking water bath
-
10-ml pipets, sterile
-
Refrigerated centrifuge, 4°C
-
100-, 70-, and 40-μm nylon mesh cell strainer filters, sterile (Falcon, cat. no. C352360, C352350, and C352340)
-
Hemocytometer (Sigma-Aldrich, Z359629, or similar)
-
Tissue culture microscope
-
Additional reagents or equipment for rat euthanasia
1.Euthanize Sprague-Dawley rats as per institutionally approved protocols.
2.Perform initial dissection of the testis and attached epididymis using sterile surgical instruments (blunt-tip fine tweezers, small fine-point scissors, and scalpel). Place the tissues immediately into a 50-ml conical tube containing pre-warmed (37°C) DMEM/Ham F12 supplemented with penicillin-streptomycin, until all of the animals have been dissected.
3.Under aseptic conditions in a laminar flow hood, transfer the tissue and a small volume of the pre-warmed (37°C) medium into a 100-mm Petri dish. Dissect the epididymis away from the testis by first grasping the tissue, using blunt-tip fine tweezers, at the junction between the testis and the epididymis and then cutting away the epididymis from the testis using small fine-point scissors or a scalpel.
4.Trim the fatty tissue away from the epididymis using a sterile scalpel blade, carefully following the epididymis along its length until all of the surrounding fatty and connective tissue has been cut away. Cut the tissue into small fragments (∼1 mm3).
5.Transfer the tissue fragments into a sterile 125-ml Erlenmeyer flask (tissue from two animals per flask) containing 25 ml pre-warmed (37°C) digestion medium. Cover with aluminum foil and Parafilm to keep sterile and incubate for 50 min at 37°C in a shaking water bath. Then, remove from water bath and return to laminar flow hood. Using a sterile 10-ml pipet, dissociate the tissue by mixing the solution up and down 5 to 6 times. Allow the tissue to sediment for 10 min, remove 10 ml medium using a sterile 10-ml pipet, and add an equal volume (∼10 ml) of fresh digestion medium with a new sterile 10-ml pipet. Once again, cover the flask with the Parafilm and aluminum foil.
6.Return flasks to water bath and incubate an additional 50 min at 37°C with agitation.
7.Remove the flasks from the water bath and return to the laminar flow hood. Once again (as in step 5), mix the cells by gently pipetting the cell solution up and down using a sterile 10-ml pipet. Allow the cells to sediment at the bottom of the flask for 10 min.
8.Remove as much of the medium as possible (∼23 ml of 25 ml) without aspirating the cells.
9.Add fresh digestion medium (10 to 15 ml), mix by swirling the flask gently, allow the cells to sediment to the bottom of the flask for 10 min, and then remove excess medium (only 2 to 3 ml medium should remain).
10.Add 10 ml of 1× trypsin-EDTA and incubate for 10 min at 37°C using a shaking water bath. Dissociate the cells by gently pipetting up and down using a sterile 10-ml pipet.
11.Add 10 ml pre-warmed (37°C) DMEM/Ham F12 supplemented with penicillin-streptomycin and 5% FBS to inhibit the trypsin activity.
12.Transfer the cell suspension from each flask into a 50-ml conical tube and collect the cells by centrifugation for 5 min at 100 × g , 4°C.
13.Return the tubes to the laminar flow hood. Carefully remove the supernatant using a sterile 10-ml pipet and resuspend the cells in 10 ml pre-warmed (37°C) DMEM/Ham F12 supplemented with penicillin-streptomycin and 5% FBS.
14.Filter the cells by successively passing them through 100-, 70-, and 40-μm nylon mesh cell strainer filters and collect the cells each time in a 50-ml conical tube. Pool the suspensions into one tube.
15.Remove a 10-μl aliquot of cells from the cell suspension and dilute 1:1 with trypan blue solution. Count the cells using a hemocytometer and a tissue culture microscope.
Basic Protocol 2: MAGNETIC ACTIVATED CELL SORTING AND ISOLATION OF BASAL CELLS
This protocol uses the total cell suspension from the Basic Protocol 1 and yields two cell suspensions: one containing primarily basal cells of the epididymis and the second containing all other epididymal cell types. The isolation of basal cells is accomplished by incubating the total cell suspension with a mouse anti-CD49f antibody, a basal cell membrane marker, followed by incubation with anti-mouse IgG microbeads. The basal cells are then isolated by magnetic cell sorting.
Materials
-
Total cell suspension from Basic Protocol 1
-
Magnetic activated cell sorting (MACS) buffer (see recipe), 4°C
-
Anti-CD49f monoclonal antibody (Bio-Rad, cat. no. MCA2034; RRID AB_2128298)
-
Anti-mouse IgG microbeads (Miltenyi Biotec; cat. no. 130-048-401 or 130-048-402)
-
Refrigerated centrifuge, 4°C
-
MACS Cell Separation MS Columns (Miltenyi Biotec, cat. no. 130-042-201)
-
Magnetic column stand (Miltenyi Biotec, cat. no. 130-042-303)
-
15-ml conical tubes, sterile
-
Additional reagents and equipment for cell counting (see Basic Protocol 1)
NOTE : All steps listed below should be done on ice unless indicated otherwise.
1.Centrifuge the total cell suspension from Basic Protocol 1 for 5 min at 300 × g , 4°C.
2.Following centrifugation, return the tube to the laminar flow hood and remove the medium by partially inverting the tube to pour off the supernatant, being careful not to dislodge the cell pellet.
3.Resuspend the cell pellet in cold MACS buffer and place the tube on ice.
4.Incubate the cells with anti-CD49f monoclonal antibody (1 μg/106 target cells) in cold MACS buffer on ice for 20 min with gentle shaking every 5 min by tapping the tube.
5.Wash the cells in 2 ml cold MACS buffer and centrifuge for 10 min at 300 × g , 4°C. Carefully decant the supernatant.
6.Repeat the wash and centrifugation from step 5 twice more, for a total of three washes and centrifugations.
7.Decant the supernatant and resuspend the cell pellet in 80 μl cold MACS buffer per 107 cells.
8.Add 20 μl anti-mouse IgG microbeads and mix gently by tapping the tube. Incubate the cells for 15 min on ice, mixing gently by tapping the bottom of the tube every 5 min.
9.Wash cells in 2 ml cold MACS buffer and centrifuge for 10 min at 300 × g , 4°C. Repeat twice more as in step 6.
10.After the third wash and centrifugation, resuspend the cell pellet in 1 ml cold MACS buffer and place the tube on ice.
11.Place the MACS Cell Separation MS Columns in a magnetic column stand. Equilibrate the columns by washing with 500 μl cold MACS buffer (this can be done during the last centrifugation step in step 9).
12.Apply the cell suspension to a column. Collect the cells in a 15-ml conical tube.
13.Rinse the column with 500 μl cold MACS buffer and discard the rinse buffer.
14.Remove the column from the magnetic stand and place a fresh 15-ml conical tube beneath the column. Apply 1 ml cold MACS buffer to the column and insert the piston (of the column) into the column. Gently apply pressure for ∼20 s to elute the cells.
15.Place the cell suspension on a second equilibrated column and repeat step 14 to purify the cell fraction a second time.
16.Following magnetic separation, count an aliquot of cells from both the positive and negative fractions (as in Basic Protocol 1, step 15).
Basic Protocol 3: PREPARATION AND CULTURE OF EPIDIDYMAL BASAL CELL ORGANOIDS
This protocol describes the 3D culture of basal cells from Basic Protocol 2 in drops composed of 1:1 Advanced DMEM/F12/Matrigel. The use of Matrigel favors 3D culture and reproduces the extracellular matrix, and a specific medium composed of several growth supplements promotes the formation and growth of organoids. Organoids appear after 3 to 4 days of culture, and their size and number can be used as a rapid means of monitoring growth.
Materials
-
Cell suspensions (positive basal cell and negative non-basal cell fractions) from Basic Protocol 2
-
Advanced DMEM/F12 (Gibco/Thermo Fisher, cat. no. 12634010), 4°C
-
Matrigel, growth factor reduced, phenol-red free (Corning, cat. no. 356231), 4°C
-
Complete basal cell organoid culture medium (Advanced DMEM/F12 containing supplements; see recipe), 32°C
-
15-ml conical tubes, sterile
-
Refrigerated centrifuge, 4°C
-
Filtered pipet tips, sterile, 4°C
-
24-well plates or 8-chamber slides (Nunc Lab-Tek II, Corning, cat. no. 154534), 37°C
-
37°C, 5% CO₂ and 32°C, 5% CO₂ cell culture incubators
-
Light microscope and/or Cytation 5 Imaging Multi-Mode Reader (BioTek)
1.After determining the cell concentration in Basic Protocol 2, step 16, take the required volumes of the cell suspensions (positive basal cell and negative non-basal cell fractions) from Basic Protocol 2 to prepare the desired numbers of drops. Transfer the cells into 15-ml conical tubes.
2.Centrifuge the cell suspensions for 5 min at 300 × g , 4°C.
3.Carefully remove most of the supernatant without disturbing the cell pellets.
4.Resuspend the cells in cold Advanced DMEM/F12 (without any supplements) using a volume of medium appropriate to achieve twice the desired concentration.
5.Add the equivalent volume of cold Matrigel to the medium using cold filtered pipet tips. Keeping the tube on ice, mix by pipetting up and down several times.
6.From this ice-cold mixture, using cold filtered pipet tips, take 50-μl aliquots to form a 50-μl drop in the center of the wells of either a pre-warmed (37°C) 24-well plate or 8-chamber slide at room temperature. Repeat until the total number of drops desired has been plated.
7.When all drops are done, wait until the drops have solidified (∼10 min), place the lid or cover on the plate or chamber slide, and rapidly flip the plate or chamber slide upside down.
8.Incubate the inverted plate or slide for 30 min at 37°C, 5% CO₂, in a cell culture incubator.
9.Following this initial incubation, remove the plate or slide from the incubator and add pre-warmed complete basal cell organoid culture medium (32°C), with 500 μl per well for a 24-well plate and 250 μl per chamber for an 8-chamber slide.
10.Culture at 32°C and 5% CO₂ in a cell culture incubator and monitor daily. After 2 to 3 days, remove half of the medium and replace with fresh complete medium. Repeat this process every 2 to 3 days.
11.Assess the formation and number of organoids over time by microscopy using a light microscope and/or Cytation 5 Imaging Multi-Mode Reader.
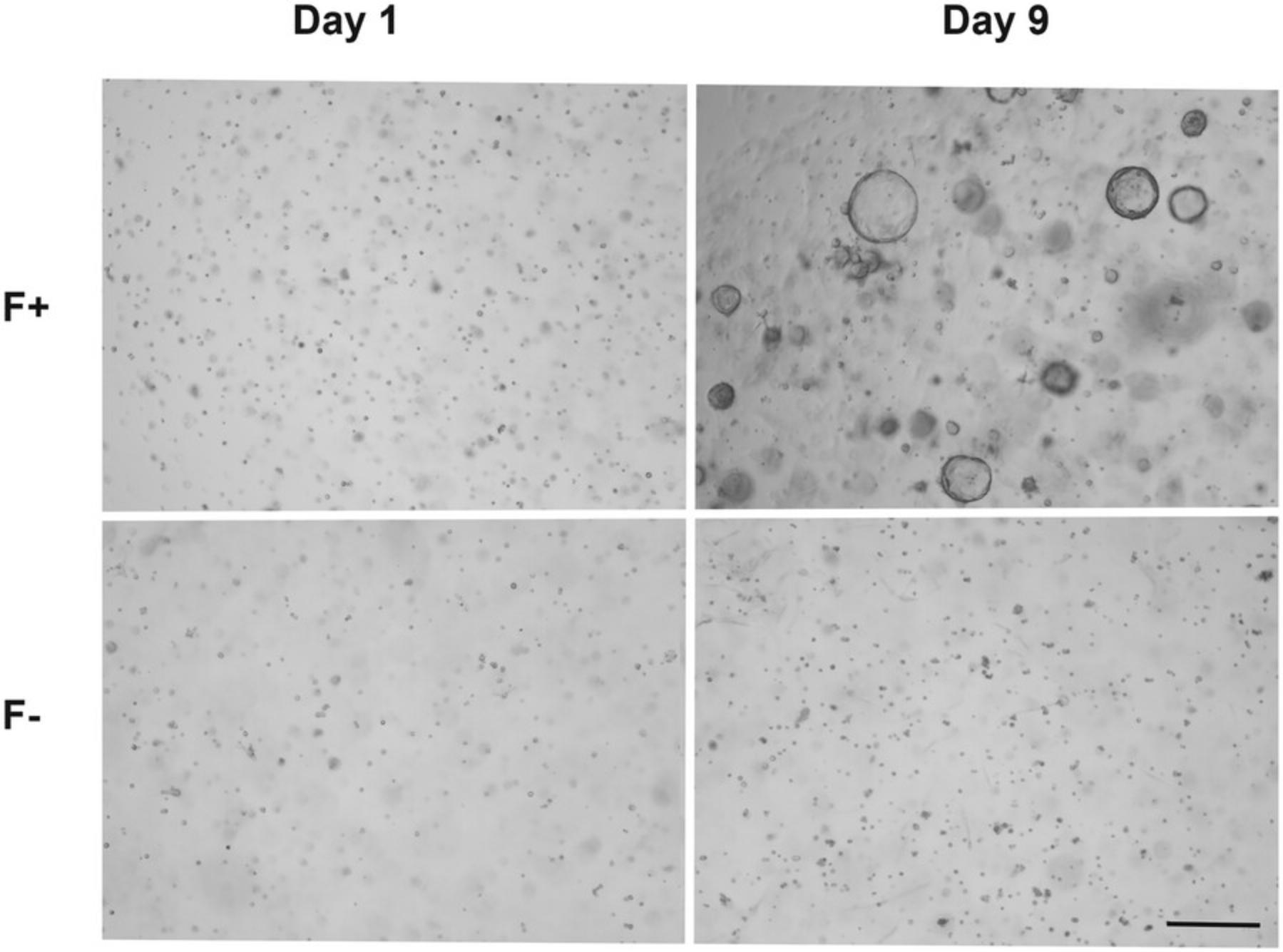
Basic Protocol 4: PASSAGE OF EPIDIDYMAL BASAL CELL ORGANOIDS
The ability of basal cells to self-renew allows them to be passaged multiple times and to preserve their ability to form new organoids over time. The passage of organoids requires several steps, which are (1) the enzymatic removal of the Matrigel with Dispase; (2) the enzymatic and mechanical dissociation of the organoids with trypsin and a syringe, respectively; (3) the formation of new drops of Matrigel with dissociated cells; and (4) the use of an adapted medium to promote the formation of new organoids.
Materials
-
Dispase II (Gibco/Thermo Fisher, cat. no. 17105041)
-
Epididymal basal cell organoids (see Basic Protocol 3)
-
1× trypsin-EDTA (Gibco/Thermo Fisher Scientific, cat. no. 15400054)
-
DMEM/Ham F12 medium supplemented with 5% (v/v) FBS (Corning, cat. no. 35-077-CV), 37°C
-
Trypan blue solution (Gibco/Thermo Fisher Scientific, cat. no. 15250061)
-
Complete basal cell organoid culture medium (see recipe) supplemented with cholera toxin, Noggin, R-spondin1, and Y-27632 dihydrochloride (see recipes; added at 1 μl per 1 ml complete medium)
-
22-µm filter and syringe
-
37°C incubator and/or water bath
-
Filtered pipet tips, sterile
-
15-ml conical tubes, sterile
-
Standard tabletop centrifuge
-
1.5-ml tubes, sterile
-
1-ml syringe with 27-G needle
-
Light microscope
-
Hemocytometer
-
Additional reagents or equipment for organoid culture (see Basic Protocol 3, steps 4 to 10)
1.Prepare 1 mg/ml Dispase II solution, sterilize by filtration using a 22-µm filter and syringe, and pre-warm at 37°C in an incubator or water bath.
2.Remove the culture medium from the epididymal basal cell organoids and incubate organoids at 37°C with 1 ml of 1 mg/ml Dispase II until complete dissociation of the Matrigel, ∼1 hr. After this time, if the drops are not liquified, use a 1000-µl pipet with a sterile filtered pipet tip and dissociate the Matrigel by pipetting up and down and then incubating at 37°C for 15 min.
3.Once the Matrigel is completely digested, pipet up and down again and collect the suspension in 15-ml conical tubes, with one tube per well or chamber. Rinse the wells or chambers with 1 ml of 1 mg/ml Dispase II to collect any remaining organoids. Pool into the same tube used for that well or chamber previously.
4.Collect the organoids by centrifugation for 7 min at 300 × g at room temperature in the 15-ml conical tubes. Remove supernatant carefully.
5.Resuspend the organoids in 250 µl of 1× trypsin-EDTA and transfer the cells into a 1.5-ml tube. Add an additional 250 µl trypsin-EDTA to the 15-ml conical tube to collect any cells left and add to the 1.5-ml tube. Incubate for 10 min at 37°C.
6.Stop trypsin activity with the addition of 500 µl pre-warmed DMEM/Ham F12 medium supplemented with 5% FBS.
7.Collect cells and organoid fragments by centrifugation for 7 min at 300 × g at room temperature. Resuspend them in 1 ml DMEM/Ham F12 medium supplemented with 5% FBS.
8.Take the resuspended cell solution and pass it through a 1 ml syringe with a 27-G needle. Repeat up to five times, until a single-cell suspension is obtained. Verify the preparation to ensure that all of the cells are dispersed into single cells by examining a drop of the suspension under a light microscope.
9.Remove a 10-µl aliquot of cells and mix 1:1 with trypan blue solution. Incubate at room temperature for 1 to 2 min and then transfer the mixture to a hemocytometer slide and count the cells under a light microscope.
10.Resuspend the cells in an appropriate volume of cell culture medium plus Matrigel at the desired concentration and dispense accordingly into fresh wells or chamber slides as described in Basic Protocol 3, steps 4 to 10, and maintain in complete basal cell organoid culture medium supplemented with cholera toxin, Noggin, R-spondin1, and Y-27632 dihydrochloride.
Basic Protocol 5: FREEZING AND THAWING OF EPIDIDYMAL BASAL CELL ORGANOIDS
This protocol describes the method for freezing dissociated cells following the passage of organoids (Basic Protocol 4), as well as the method of thawing of these cells for subsequent 3D culture and formation of new organoids (Basic Protocol 3).
Materials
-
Dissociated cells (see Basic Protocol 4)
-
CryoStor® CS10 Cell Freezing Medium (Stem Cell Technologies, cat. no. 07930), 4°C
-
70% (v/v) ethanol
-
DMEM/Ham F12 with 1% (w/v) bovine serum albumin (BSA; low endotoxin; Bio Basic, cat. no. AR2442) (filtered, sterile), 37°C
-
Advanced DMEM/F12 (Gibco/Thermo Fisher, cat. no. 12634010), 37°C
-
Standard tabletop centrifuge
-
2-ml cryotubes, sterile
-
Cooler (CoolCell freezing system, Corning, cat. no. 432000)
-
−150°C freezer or liquid nitrogen storage tank
-
37°C water bath
-
15-ml conical tubes, sterile
-
Additional reagents and equipment for cell counting (see Basic Protocol 4, step 9) and organoid culture (see Basic Protocol 4, step 10)
1.After counting (see Basic Protocol 4, step 9), centrifuge the dissociated cells for 7 min at 300 × g at room temperature.
2.Under aseptic conditions, remove the supernatant carefully and add cold CryoStor® CS10 Cell Freezing Medium to obtain the adequate concentration.
3.Transfer the solution into sterile 2-ml cryotubes under aseptic conditions.
4.Place the tubes in a cooler at −80°C for a few days at most.
5.Transfer to −150°C freezer or preferably the vapor phase of liquid nitrogen for long-term storage.
6.To thaw cells to use to form new organoids, thaw a cryotube in a 37°C water bath for ∼2 min or until the solution is almost totally thawed.
7.Spray and wipe the exterior of the cryotube with 70% ethanol and transfer to laminar flow hood.
8.Add 1 ml pre-warmed DMEM/Ham F12 with 1% BSA to the cryotube.
9.Transfer the entire contents of the tube (∼2 ml) to a 15-ml conical tube containing 2 ml pre-warmed DMEM/Ham F12 with 1% BSA.
10.Centrifuge 7 min at 300 × g at room temperature.
11.Resuspend the pellet in Advanced DMEM/F12 and do a cell count, as described previously (see Basic Protocol 4, step 9).
12.Resuspend the cells in an appropriate volume of cold Advanced DMEM/F12 medium plus Matrigel at the desired concentration, dispense accordingly into fresh wells or chamber slides, and maintain in complete basal cell organoid culture medium with supplements (see Basic Protocol 4, step 10).
Basic Protocol 6: IMMUNOFLUORESCENT STAINING OF EPIDIDYMAL BASAL CELL ORGANOIDS
Organoid staining can be performed directly in chamber slides but can also be performed by transferring the organoids into paraffin or cryo-blocks. We describe here a protocol in which we fix organoids in a chamber slide using methanol to preserve the Matrigel drop and the 3D structure of the organoids. This permits visualization of the entire organoid by creating z-stacks of images using a confocal microscope. The washing steps and antibody incubation times are longer than in the standard immunofluorescent staining protocol in order to allow the solutions to pass through the Matrigel. The main steps of this protocol are fixation with methanol, permeabilization with Triton, blocking with BSA ± serum, primary and secondary antibody incubation, and mounting.
Materials
-
Phosphate-buffered saline (PBS)
-
Methanol (store in small bottle in −20°C freezer), −20°C
-
PBS-glycine: 0.75% (w/v) glycine (Bio Basic, cat. no. GB0235) in PBS, 37°C
-
0.03% (v/v) Triton X-100 in PBS (Fisher BioReagents, cat. no. BP151100)
-
Blocking solution: 5% (w/v) BSA (Bio Basic, cat. no. AD0023) in PBS-glycine, 5% (v/v) normal serum (species of host secondary antibody), or combination of 5% (v/v) normal serum plus 3% to 5% (w/v) BSA (Bio Basic, cat. no. AD0023) in PBS-glycine
-
Primary antibody
-
Fluorescent-labeled secondary antibody
-
5 μg/ml Hoechst 33342 dye (Biotium, cat. no. 40046)
-
Mounting medium (e.g., Fluoromount-G, SouthernBiotech, cat. no. 0100-01)
-
Parafilm
-
Coverslip
-
Blotting paper
-
Confocal microscope
1.Grow organoids, as described in Basic Protocols 3 and 4, in Nunc Lab-Tek II chamber slides (with 8 chambers).
2.Carefully remove the medium and rinse organoid culture drops with 300 to 400 μl PBS.
3.Add ice-cold methanol (pre-cooled to −20°C) slowly, drop by drop, into a corner of each chamber until the organoid culture drop is covered. Fix for 20 min at −20°C.
4.Remove methanol and rinse three times in warm PBS-glycine for 10 min each at room temperature.
5.Following the PBS-glycine washes, permeabilize the organoids in 0.03% Triton X-100 in PBS for 15 min at room temperature.
6.Wash twice with PBS-glycine for 5 min each.
7.Block with blocking solution for 90 min at room temperature.
8.Gently remove the blocking solution and replace with 200 μl of a solution of primary antibody diluted with blocking solution to the appropriate concentration. Wrap the chambered slide plus slide cover in Parafilm. Perform primary antibody incubation overnight (∼18 hr) at 4°C.
9.Following primary antibody incubation, place the chambered slide at room temperature for 30 min to solidify the Matrigel. Then, gently remove the primary antibody solution with a pipet (using a fresh tip for each chamber of the slide). Proceed immediately to next step to ensure that each chamber containing organoids does not dry out.
10.Wash three times in PBS-glycine, added dropwise, for 10 min each.
11.Incubate with the appropriate fluorescent-labeled secondary antibody diluted in blocking solution containing Hoechst 33342 dye (5 μg/ml solution diluted 1:2000, v/v) for 90 to 120 min in the dark at room temperature.
12.Wash three times in PBS-glycine for 10 min each.
13.Carefully remove any excess PBS-glycine from each chamber. Gently remove the chamber divisions from the slide, taking care to avoid touching or disrupting the organoids.
14.Add mounting medium such as Fluoromount-G by adding a small drop (∼25 μl) to each chamber and then cover the entire slide with a coverslip. Blot excess mounting medium using blotting paper.
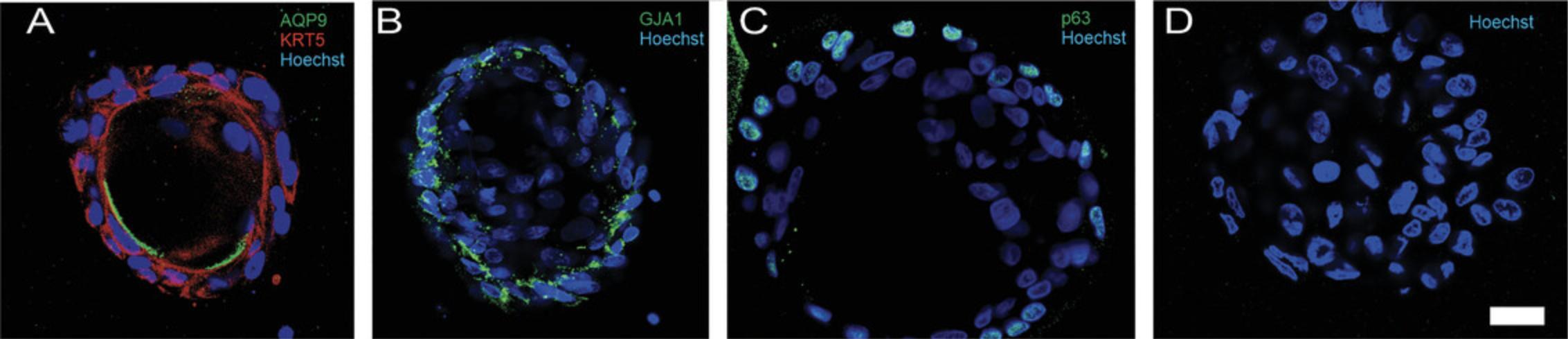
15.Assess the organoids by immunofluorescence using a confocal microscope (Fig. 3).
REAGENTS AND SOLUTIONS
A8301, 100 μM
Dissolve 5 mg A8301 (MilliporeSigma, cat. no. SML0788) in 11.86 ml dimethyl sulfoxide (DMSO) (1 mM stock solution I). Add 100 μl of this stock solution to 900 μl DMSO in a sterile 1.5-ml tube and mix by vortexing (100 μM stock solution II). Dispense stock solution I into aliquots of 1 ml and stock solution II into aliquots of 50 μl in sterile 1.5-ml tubes and store ≤3 months at −20°C.
Cholera toxin, 1 mg/ml
Dissolve 0.5 mg cholera toxin (MilliporeSigma, cat. no. C8052) in 0.5 ml sterile ddH2O. Spin the tube before opening. Dispense into aliquots of 10 μl in sterile 1.5-ml tubes and store ≤ 3 months at 4°C. Use 0.1 μl per 1 ml complete medium.
Complete basal cell organoid culture medium
Combine all of the components listed in Table 1 in a sterile 50-ml conical tube. Filter through a 0.2-μm filter and store ≤1 month at 4°C.
| Component |
Stock concentration |
Final concentration |
For 50 ml medium |
|---|---|---|---|
| Advanced DMEM/F12 (Gibco/Thermo Fisher, cat. no. 12634010) | - | - | 47.501 ml |
| A8301 (see recipe) | 100 μM | 250 nM | 125 μl |
| cAMP (see recipe) | 100 μg/ml | 10 ng/ml | 20 μl |
| DHT (see recipe) | 100 μM | 10 nM | 5 μl |
| GlutaMAX (see recipe) | 100× | 1× | 500 μl |
| Heparin (see recipe) | 500 μg/ml | 4 μg/ml | 400 μl |
| HEPES (see recipe) | 1 M | 100 mM | 500 μl |
| EGF (see recipe) | 10 μg/ml | 10 ng/ml | 50 μl |
| bFGF (see recipe) | 10 μg/ml | 10 ng/ml | 50 μl |
| Hydrocortisone (see recipe) | 1 mg/ml | 80 ng/ml | 4 μl |
| NAC (see recipe) | 500 mM | 1.25 mM | 125 μl |
| Vitamin E (see recipe) | 20 μg/ml | 200 ng/ml | 500 μl |
Cyclic adenosine monophosphate (cAMP), 100 μg/ml
Dissolve 1 mg cAMP (MilliporeSigma, cat. no. A9501) in 1.0 ml sterile ddH2O in a sterile 1.5-ml tube by vortexing (1 mg/ml stock solution I). Add 100 μl of this stock solution to 900 μl sterile ddH2O in a sterile 1.5-ml tube and mix by vortexing (100 μg/ml stock solution II). Dispense stock solution I into aliquots of 100 μl and stock solution II into aliquots of 10 or 20 μl in sterile 1.5-ml tubes and store ≤3 months at −20°C.
Dihydrotestosterone (DHT), 100 μM
Dissolve 29.04 mg DHT (MilliporeSigma, cat. no. A8280) in 1.0 ml anhydrous ethanol in a sterile glass vial by vortexing (100 mM stock solution I). Add 100 μl of this stock solution to 900 μl of anhydrous ethanol in a sterile glass vial (Fisherbrand, cat. no. FS60940D1 or FS60940HD12) and mix by vortexing (10 mM stock solution II). Add 100 μl of this stock solution to 900 μl anhydrous ethanol in a sterile glass vial and mix by vortexing (1 mM stock solution III). Again, add 100 μl of this stock solution to 900 μl of anhydrous ethanol in a sterile glass vial and mix by vortexing (100 μM stock solution IV). Use Parafilm to tighten the caps of the vials and avoid any evaporation of the solutions. Store solutions I to IV ≤3 months at −20°C.
GlutaMAX, 100×
Store GlutaMAX (Gibco/Thermo Fisher, cat. no. 35050061) ≤3 months at room temperature. It can be aliquoted into sterile 15-ml conical tubes for convenience.
Heparin, 500 μg/ml
Prepare a stock solution by dissolving 5 mg heparin (MilliporeSigma, cat. no. H3149) in 10 ml ddH2O. Filter-sterilize by passing through 0.22-μm syringe filter into a sterile 15-ml conical tube. Store ≤3 months at 4°C.
HEPES, 1 M
Prepare a stock solution by dissolving 23.83 g HEPES (Gibco/Thermo Fisher, cat. no. 15630080) in 100 ml ddH2O. Adjust the pH to 7.4. Filter-sterilize by passing through 0.22-μm syringe filter and aliquot into 10 sterile 15-ml conical tubes. Store ≤3 months at −20°C and keep an aliquot at 4°C.
Human basic fibroblast growth factor (bFGF), 10 μg/ml
Take up 0.5 ml sterile ddH2O into a 1-ml syringe and inject into a vial of lyophilized bFGF (50 μg; Sino Biological, cat. no. 10014-HNAE). Swirl to dissolve and remove entire solution with syringe. Transfer to a sterile 15-ml conical tube. Rinse the vial with an additional 0.5 ml sterile ddH2O and add to the 15-ml tube. Complete volume to 5 ml with sterile ddH2O. Prepare aliquots (100 or 200 μl) in sterile 1.5-ml tubes and store ≤3 months at −20°C.
Human epidermal growth factor (EGF), 10 μg/ml
Take up 2 ml sterile PBS into a 3-ml syringe and inject into a vial of lyophilized EGF (100 μg; Gibco/Thermo Fisher, cat. no. PHG0311). Swirl to dissolve and remove entire solution with syringe. Transfer to a sterile 15-ml conical tube. Rinse the vial with an additional 2 ml sterile PBS and add to the 15-ml tube. Complete volume to 10 ml with sterile PBS. Prepare aliquots (100 or 200 μl) in sterile 1.5-ml tubes and store ≤3 months at −20°C.
Hydrocortisone, 1 mg/ml
Dissolve 1 mg hydrocortisone (MilliporeSigma, cat. no. H0888) in 1 ml anhydrous ethanol. Vortex and dispense into aliquots (50 μl) in sterile 1.5-ml tubes. Store ≤3 months at −20°C.
Magnetic activated cell sorting (MACS) buffer
- 2 mM EDTA
- 0.5% (w/v) BSA (Bio Basic, cat. no. AD0023)
- PBS
- Adjust to pH 7.2
- Filtered through 0.2-μm filter
- Prepare 1 day before use and store at 4°C
N-acetyl cysteine (NAC), 500 mM
Dissolve 0.408 g NAC (MilliporeSigma, cat. no. A9165) in 5 ml sterile ddH2O. Vortex well and dispense into aliquots (100 or 200 μl) in sterile 1.5-ml tubes. Store ≤3 months at −20°C.
Noggin, 100 μg/ml
Centrifuge the vial of Noggin (PeproTech, cat. no. 120-10C) before opening. Add 200 μl filtered sterilized 1× PBS containing 0.1% (w/v) BSA (Bio Basic, cat. no. AR2442) to 20 μg Noggin. Mix by pipetting up and down. Do not vortex. Dispense into aliquots of 50 μl in sterile 1.5-ml tubes and store ≤ 3 months at −20°C. Use 1 μl per 1 ml complete medium.
R-spondin1, 500 μg/ml
Centrifuge the vial of R-spondin1 (PeproTech, cat. no. 120-38) before opening. Add 200 μl filtered sterilized 1× PBS containing 0.1% (w/v) BSA (Bio Basic, cat. no. AR2442) to 100 μg R-spondin1. Mix by pipetting up and down. Do not vortex. Dispense into aliquots of 50 μl in sterile 1.5-ml tubes and store ≤ 3 months at −20°C. Use 1 μl per 1 ml complete medium.
Vitamin E (tocopherol), 20 μg/ml
Dissolve 21 μl vitamin E (MilliporeSigma, cat. no. T1539) in 1.0 ml anhydrous ethanol in a sterile 1.5-ml tube by vortexing (20 mg/ml stock solution I). Add 10 μl of this stock solution to 9.99 ml anhydrous ethanol in a sterile 15-ml conical tube and mix well by vortexing (20 μg/ml stock solution II). Dispense the stock solution II into aliquots (1 ml) in sterile 1.5-ml tubes. Use Parafilm to tighten the caps of the 1.5-ml tubes to avoid any evaporation of the solutions. Store the stock solutions I and II ≤3 months at −20°C.
Y-27632 dihydrochloride, 100 mM
Add 300 μl of filtered sterilized PBS 1× to 10 mg of Y-27632 dihydrochloride (BioGems, cat. no. 1293823). Mix by pipetting up and down. Do not vortex. Dispense into aliquots of 5 μl in sterile 1.5-ml tubes and store ≤ 3 months at −20°C. Use 0.1 μl per 1 ml complete medium.
COMMENTARY
Background Information
Given that the epididymal epithelium primarily comprises principal cells and that basal cells account for only 10% to 15% of the total number of epithelial cells in the epididymis, many of the studies on the epididymis have focused on principal cells. The role of basal cells in the epididymis has been a source of controversy. Although initial studies suggested that these cells were epididymal stem cells, studies in aging rodents suggested that the limited proliferation of epididymal cells was not conducive to the presence of stem cells (Clermont & Flannery, 1970). However, these studies were done in aging animals, in which the epididymal epithelium is known to have a particularly low rate of cell proliferation (Pinel et al., 2019). Furthermore, studies identified quiescent stem cells in other tissues where there existed a low rate of proliferation, such as liver, kidney, and trachea (Huch et al., 2013; Kim et al., 2012; Li & Clevers, 2010). These stem cells, referred to as adult, or mesenchymal, stem cells, lay dormant until activated by tissue damage or cell death (Barriga et al., 2017; Bartfeld & Clevers, 2017; Sangiorgi & Capecchi, 2008; Yan et al., 2012). A characteristic of quiescent stem cells is that they express a number of proteins and enzymes implicated in the metabolism of free radicals, which is necessary to favor the long-term survival of these cells. It is therefore perhaps not surprising that among the first markers of epididymal basal cells were superoxide dismutase, various glutathione reductases, and metallothionein, which all play a role in the reduction of oxidative stress (Cheung et al., 2005; Cyr et al., 2001; Papp et al., 1995; Veri et al., 1993).
Epididymal basal cells are characterized by having thin long projections that can reach the lumen of the epididymis (Martan et al., 1964; Shum et al., 2008). Interestingly, these projections are prominent in the initial segment of the murine epididymis, but in the rat and human, these projections are more prominent in the corpus and cauda epididymidis (Mandon & Cyr, 2015). It has been suggested that the apical region of these projections may play a role as sensors of luminal pH (Shum et al., 2008; Shum et al., 2013; Shum et al., 2011). Furthermore, these basal cell projections express the tight junction protein claudin 1 (Cldn1) and appear to cross the BEB (Gregory et al., 2001; Shum et al., 2008).
There were no commercially available epididymal basal cell lines, so in order to fully understand the role(s) of basal cells, we developed a protocol to isolate basal cells from the rat epididymis using magnetic bead separation and anti-CD49f antibody. We were able to consistently achieve >90% purity in our preparations (Mandon et al., 2015). Using transcriptomic analyses, we showed that basal cells shared common properties with adult stem cells. Furthermore, we showed that in 2D culture, basal cells could differentiate into cells that expressed markers similar to those of epididymal principal cells. Subsequent studies by Mou et al. (2016) showed that cell grafts of KRT5+ murine basal cells implanted under the skin could differentiate into spheroid structures containing both basal cells and clear cells, indicating that these cells were in fact stem cells. Further studies showed that basal cells are derived from columnar cells, a stem cell population present in the epididymis at birth (Dufresne et al., 2022).
Adult stem cells are characterized by having the ability to form organoids, to self-renew, and to differentiate in vitro. Our lab showed that isolated basal cells could form organoids in vitro using a 3D cell culture approach and that cells from these organoids had the ability to self-renew (Pinel & Cyr, 2021). In fact, the cells could be maintained in culture for >2 months without any obvious decrease in ability to self-renew (Pinel & Cyr, 2021). Finally, we also showed that basal cells from these organoids could differentiate into principal cells, regardless of the region along the epididymis from which the basal cells had been isolated. The principal cells that were differentiated from basal cell organoids not only were morphologically similar to principal cells from tissue but also appeared to secrete clusterin, a protein that is secreted by principal cells that binds to developing sperm in the epididymis (Pinel & Cyr, 2021). This demonstrated that these differentiated principal cells were functional.
The rat 3D epididymal basal cell organoid model offers a novel tool for studies regarding effects of a substance on this region of the male reproductive tract. Although it is not a complete system as in whole-animal in vivo studies, it does provide an opportunity to examine specific effects (of a substance) on the epididymal epithelium. Furthermore, it permits evaluation of any potential impacts on growth and epithelial cell differentiation and a better understanding of mechanism(s) of action of a substance, thereby providing insight regarding more accurate therapeutics, for example. Finally, it allows for a significant reduction in the use of whole animals and the associated costs.
A significant advantage is the fact that cell-cell interactions in a 3D environment are achieved in an organoid system, via the development and differentiation of stem cells into various types of mature epithelial cells. This represents an important advancement in the current limited understanding of epididymal cellular differentiation, which is a lengthy process in the epididymis, as well as understanding of epithelial regeneration and the factors that regulate both differentiation and regeneration. As such, having an in vitro model represents an important system that can be exploited to tease out the factors involved in the differentiation of basal cells. The fact that these cells can be genetically manipulated allows for future studies to examine which signaling pathways play a role in cell proliferation and differentiation. All of these can be subsequently validated using transgenic approaches. With respect to toxicology, in vitro models offer high-throughput approaches to understand mechanisms of toxicity and to predict effects. This 3D system does provide a unique opportunity to test a substance, either as a toxicant or as a therapeutic agent, in a relatively rapid, more uniform, and controlled system and offers the possibility of addressing more specific mechanistic or molecular aspects of toxicology or biomedical studies.
Although there are numerous advantages to organoid models, there are limitations that have yet to be overcome. The process of generating rat 3D epididymal basal cell organoids is time consuming, fairly complex, and somewhat costly. These factors apply to other organoid models as well. In addition to the aforementioned time and cost aspects, similar to nearly all in vitro systems, there is no vascular, nervous, or immune system as in the whole organism. Hence, the influence of these systems and their interactions with the model cells are lacking. Furthermore, because organoids are derived from single stem cells, their interactions with the immune, vascular, and nervous systems are lacking. The immune and vascular systems may be particularly important in the epididymis, where there is increasing evidence that these may influence cellular differentiation. Changes in regional function along the epididymis and differences in the composition of luminal factors versus blood and their contributions to epididymal physiology also need to be considered. Some of these aspects are being developed in other labs with other tissue organoid systems but have yet to be well established. Consequently, these limitations must be considered when interpreting experimental observations and data.
In addition to the rat organoid model, studies have also established organoids and spheroid models using cells from both the mouse and human epididymides (Kristensen et al., 2010; Leir et al., 2020; Mou et al., 2016). These have been discussed in detail in a recent review (Cyr & Pinel, 2022).
There are currently no other published protocols for generation of rat epididymal organoids. The development of rat epididymal organoids, particularly those derived from basal cells, as described here, represents a significant advancement in tools and techniques for studying epididymal function, for elucidating the role(s) of basal cells within the epididymis, and for performing toxicological studies on the epididymis.
Critical Parameters
We chose to use 48-day-old rats for these protocols because at this age, the epididymis is fully differentiated, but the spermatozoa are not yet present in the tubule, minimizing the chances of contamination with sperm during the tissue digestion. However, the addition of DNase with the collagenase (type I) can prevent this if older animals are used.
These protocols are lengthy and time consuming, so careful advance planning and preparation of materials are requisite. Experience with aseptic technique and cell culture is critical for successful execution of these protocols. Careful manipulation with respect to pipetting and changing medium is necessary in order to preserve the integrity of the cells and organoids and to prevent damage and cell death. Regular and careful observation and proper maintenance of the cell and organoid cultures are also essential. As such, experience with microscopy and with immunofluorescent staining techniques is also important. These attributes are required for correct identification and validation of basal cells and of other cell types that develop subsequently within the organoids.
Familiarity with male rat anatomy and precise dissection is also necessary. Careful dissection within a short time period improves chances of success. The testis and epididymis can be removed together in the animal facility under aseptic conditions, placed in warm culture medium, and then transported to the laboratory/laminar flow hood for subsequent dissection and manipulation. Alternatively, if one is sufficiently experienced and can readily dissect out the epididymis from the animal as an initial step, this may improve chances of success. However, because time is of the essence and the epididymis is surrounded by a considerable amount of fatty tissue in the adult, we recommend initial dissection of the testis and epididymis together, followed by a second step of dissection under the laminar flow hood. In this second step, the epididymis can be freed of the surrounding fatty and connective tissue, as well as the testis (which can be frozen or fixed for other purposes if desired).
The use of Matrigel is a critical parameter for obtaining a 3D culture. Matrigel should be aliquoted in undiluted form into small quantities and should not be frozen after thawing (the remainder of thawed aliquots can be stored at 4°C for up to 4 weeks). Matrigel should always be handled on ice and pipetted with cold tips, as it solidifies with increasing temperature.
Troubleshooting
Tables 2, 3, and 4 list common problems, their causes, and potential solutions for the protocols.
| Problem | Possible cause(s) | Potential solution(s) |
|---|---|---|
| Low cell yield | Improper dissection | Ensure that epididymis has not been damaged during dissection; trim off all surrounding tissue |
| Poor cell separation technique | Save all supernatants; repeat cell counts for all supernatants | |
| Incorrect cell count | Count more than one aliquot; verify counting method |
| Problem | Possible cause(s) | Potential solution(s) |
|---|---|---|
| Contamination | Poor aseptic technique | Sterility and aseptic technique must be maintained during all steps |
| Poor or low growth | Incorrect dilution of cells | Verify calculations and modify dilutions accordingly |
| Insufficient number of cells seeded per well or chamber | Ensure that all medium components are sufficiently fresh (see Reagents and Solutions for storage conditions and times) | |
| Matrigel drop will not hold | The 1:1 ratio has not been maintained, and there is more medium than Matrigel | Be sure to remove as much medium as possible in step 3 of Basic Protocol 3 |
| The plate was turned over too early in step 7 of Basic Protocol 3 | Wait ≥10 min for the Matrigel to start solidifying and re-turn the plate quickly | |
| Organoid growth in F- (negative) fraction | Improper or inadequate cell separation (Basic Protocol 2) | Ensure that all centrifugation steps and removal of supernatant are done carefully; check cell counts for all supernatants and fractions |
| Organoids do not grow after passaging | The supplements in the medium are not optimal | Ensure that supplements are <6 months old; make sure to add Y-27632, cholera toxin, R-spondin1, and Noggin to the medium after passaging |
| Problem | Possible cause(s) | Potential solution(s) |
|---|---|---|
| Matrigel drop will not hold | The methanol was added too quickly during the fixation step of immunofluorescence staining | Add the methanol drop by drop using a small pipet tip |
| The addition of solution was done to quickly or not in the corner of the chamber slides | Add the solution slowly and make sure to not touch the drop with the pipet | |
| Poor immunofluorescence results | Washing away of or damage to organoids during staining protocol | Add and remove any solutions gently by placing pipet tip along edge of side of each chamber of 8-chamber slide); change tips for each step and each chamber |
Understanding Results
Isolation of basal cells
The protocol to isolate basal cells from rat epididymal cells (Basic Protocol 2) was developed in the lab in 2015 and has since been used numerous times by several students and research personnel. A thorough analysis of the basal cell fraction and a comparison with the non-basal-cell fraction was conducted to confirm the efficiency of this protocol. This protocol should yield a basal cell fraction with purity ≥90% and viability >80%. Purity can be evaluated easily by taking an aliquot of cells after the basal cell isolation and preparing a smear on a glass slide that will be stained with another basal cell marker (e.g., cytokeratin 5). Low purity or viability of the basal cell fraction at this stage could indicate that a problem occurred during sorting and should prompt further investigation of the protocol, as well as hesitation to proceed to any further steps or analyses.
Organoid culture
The organoid culture protocol (Basic Protocol 3) should result in the formation of organoids after 3 to 4 days of culture of the basal cell (positive) fraction in the appropriate medium. Monitoring of the basal cells should be done regularly, and several parameters, including size and number, can be easily determined (Fig. 2). Because our protocol was one of the first to demonstrate the ability of these cells to form organoids, we also added the monitoring of specific cells to demonstrate their ability to proliferate to form organoids and not simply cell aggregates (Pinel & Cyr, 2021). We believe this specific monitoring should be done at least one or two times during the first few experiments to confirm the origin of organoids. After about 12 to 13 days of culture, we observed the first signs of degradation of the organoids, probably due to the deterioration of the Matrigel, thus justifying the need to passage them every 10 days.
The medium was optimized to favor the proliferation of basal cells during the first week of culture and the differentiation of the cells few days later, when the principal cell marker Aquaporin 9 appeared at 11 days (Fig. 3). It is, however, important to mention that all organoids will not display the same degree of differentiation at the same time. Hence, we think it is important to include quantitative data with RNA analysis or protein analysis, in addition to fluorescence staining, to determine differentiation. Other markers of principal cells or other epididymal cell types can also be used to evaluate the composition of organoids at various time points. It is also important to note that not all basal cells will develop into organoids, but these cells are still present in the Matrigel drop and will therefore be included in RNA or protein extraction.
We were able to passage basal cell organoids (Basic Protocol 4) for more than 10 generations, confirming the ability of basal cells to self-renew (Pinel & Cyr, 2021). Even though we did not observe any difference in the number or morphology of organoids between passages, we did not obtain any data regarding their genetic or chromosomal stability. As is usually done with cell lines, we recommend freezing a stock of cells (Basic Protocol 6) with a low passage number.
Time Considerations
Rats are normally acclimated to the animal holding facility for 5 to 7 days prior to use in these protocols. Basic Protocols 1 through 3 take ∼12 hr for two people to complete and must be done sequentially on the same day. There are really not any steps at which these protocols can be paused for any length of time. Basic Protocols 4 and 5 (passaging, freezing, and thawing of organoids) take approximately 1 to 2 hr. Basic Protocol 6 (immunofluorescent staining) takes approximately 7 to 8 hr in total, but this may be divided into three parts: (i) 3 to 3.5 hr up to the primary antibody step, (ii) overnight incubation with the primary antibody (18 hr), and (iii) incubation with the secondary antibody and mounting. The entire protocol can be performed within 1 day, with primary antibody incubation of 2 to 3 hr, but we recommend performing overnight incubation at 4°C.
Acknowledgments
The contribution of Marion Mandon to the development of the basal cell isolation protocol is gratefully acknowledged. L.P. received a studentship from the Réseau Québécois en Reproduction (RQR-FRQNT). The development of these protocols was funded in part by grants from CIHR (84576), NSERC (06432), and a Canada Research Chair awarded to D.G.C.
Author Contributions
Julie Dufresne : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; validation; visualization; writing—original draft; writing—review and editing. Mary Gregory : Conceptualization; data curation; formal analysis; investigation; methodology; project administration; resources; validation; visualization; writing—original draft; writing—review and editing. Laurie Pinel : Conceptualization; data curation; formal analysis; investigation; methodology; resources; validation; visualization; writing—original draft; writing—review and editing. Daniel G. Cyr : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; validation; writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data, tools, and materials (or their source) that support these protocols are available from the corresponding author upon request.
Literature Cited
- Adam, C., & Cyr, D. G. (2016). Role of specificity protein-1 and activating protein-2 transcription factors in the regulation of the gap junction protein beta-2 gene in the epididymis of the rat. Biology of Reproduction , 94(6), 120. https://doi.org/10.1095/biolreprod.115.133702
- Adamali, H. I., & Hermo, L. (1996). Apical and narrow cells are distinct cell types differing in their structure, distribution, and functions in the adult rat epididymis. Journal of Andrology , 17(3), 208–222. https://doi.org/10.1002/j.1939-4640.1996.tb01776.x
- Agarwal, A., & Hoffer, A. P. (1989). Ultrastructural studies on the development of the blood-epididymis barrier in immature rats. Journal of Andrology , 10(6), 425–431. https://doi.org/10.1002/j.1939-4640.1989.tb00132.x
- Araki, Y., Suzuki, K., Matusik, R. J., Obinata, M., & Orgebin-Crist, M. C. (2002). Immortalized epididymal cell lines from transgenic mice overexpressing temperature-sensitive simian virus 40 large T-antigen gene. Journal of Andrology , 23(6), 854–869. https://doi.org/10.1002/j.1939-4640.2002.tb02344.x
- Barrachina, F., Battistone, M. A., Castillo, J., Mallofre, C., Jodar, M., Breton, S., & Oliva, R. (2022). Sperm acquire epididymis-derived proteins through epididymosomes. Human Reproduction , 37(4), 651–668. https://doi.org/10.1093/humrep/deac015
- Barriga, F. M., Montagni, E., Mana, M., Mendez-Lago, M., Hernando-Momblona, X., Sevillano, M., Guillaumet-Adkins, A., Rodriguez-Esteban, G., Buczacki, S. J. A., Gut, M., Heyn, H., Winton, D. J., Yilmaz, O. H., Attolini, C. S., Gut, I., & Batlle, E. (2017). Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell , 20(6), 801–816.e807. https://doi.org/10.1016/j.stem.2017.02.007
- Bartfeld, S., & Clevers, H. (2017). Stem cell-derived organoids and their application for medical research and patient treatment. Journal of Molecular Medicine , 95(7), 729–738. https://doi.org/10.1007/s00109-017-1531-7
- Battistone, M. A., Elizagaray, M. L., Barrachina, F., Ottino, K., Mendelsohn, A. C., & Breton, S. (2023). Immunoregulatory mechanisms between epithelial clear cells and mononuclear phagocytes in the epididymis. Andrology , https://doi.org/10.1111/andr.13509
- Battistone, M. A., Mendelsohn, A. C., Spallanzani, R. G., Brown, D., Nair, A. V., & Breton, S. (2020). Region-specific transcriptomic and functional signatures of mononuclear phagocytes in the epididymis. Molecular Human Reproduction , 26(1), 14–29. https://doi.org/10.1093/molehr/gaz059
- Bedford, J. M., Calvin, H., & Cooper, G. W. (1973). The maturation of spermatozoa in the human epididymis. Journal of Reproduction and Fertility. Supplement , 18, 199–213.
- Breton, S., & Brown, D. (2013). Regulation of luminal acidification by the V-ATPase. Physiology , 28(5), 318–329. https://doi.org/10.1152/physiol.00007.2013
- Breton, S., Nair, A. V., & Battistone, M. A. (2019). Epithelial dynamics in the epididymis: Role in the maturation, protection, and storage of spermatozoa. Andrology , 7(5), 631–643. https://doi.org/10.1111/andr.12632
- Breton, S., Smith, P. J., Lui, B., & Brown, D. (1996). Acidification of the male reproductive tract by a proton pumping (H+)-ATPase. Nature Medicine , 2(4), 470–472. https://doi.org/10.1038/nm0496-470
- Cheung, K. H., Leung, G. P., Leung, M. C., Shum, W. W., Zhou, W. L., & Wong, P. Y. (2005). Cell-cell interaction underlies formation of fluid in the male reproductive tract of the rat. The Journal of General Physiology , 125(5), 443–454. https://doi.org/10.1085/jgp.200409205
- Clermont, Y., & Flannery, J. (1970). Mitotic activity in the epithelium of the epididymis in young and old adult rats. Biology of Reproduction , 3(3), 283–292. https://doi.org/10.1093/biolreprod/3.3.283
- Cornwall, G. A. (2009). New insights into epididymal biology and function. Human Reproduction Update , 15(2), 213–227. https://doi.org/10.1093/humupd/dmn055
- Cyr, D. G., Dufresne, J., & Gregory, M. (2018). Cellular junctions in the epididymis, a critical parameter for understanding male reproductive toxicology. Reproductive Toxicology , 81, 207–219. https://doi.org/10.1016/j.reprotox.2018.08.013
- Cyr, D. G., Dufresne, J., Pillet, S., Alfieri, T. J., & Hermo, L. (2001). Expression and regulation of metallothioneins in the rat epididymis. Journal of Andrology , 22(1), 124–135. https://doi.org/10.1002/j.1939-4640.2001.tb02162.x
- Cyr, D. G., & Pinel, L. (2022). Emerging organoid models to study the epididymis in male reproductive toxicology. Reproductive Toxicology , 112, 88–99. https://doi.org/10.1016/j.reprotox.2022.07.001
- Dube, E., & Cyr, D. G. (2012). The blood-epididymis barrier and human male fertility. Advances in Experimental Medicine and Biology , 763, 218–236. https://doi.org/10.1007/978-1-4614-4711-5_11
- Dube, E., Dufresne, J., Chan, P. T., Hermo, L., & Cyr, D. G. (2010). Assessing the role of claudins in maintaining the integrity of epididymal tight junctions using novel human epididymal cell lines. Biology of Reproduction , 82(6), 1119–1128. https://doi.org/10.1095/biolreprod.109.083196
- Dube, E., Hermo, L., Chan, P. T., & Cyr, D. G. (2008). Alterations in gene expression in the caput epididymides of nonobstructive azoospermic men. Biology of Reproduction , 78(2), 342–351. https://doi.org/10.1095/biolreprod.107.062760
- Dube, E., Hermo, L., Chan, P. T., & Cyr, D. G. (2010). Alterations in the human blood-epididymis barrier in obstructive azoospermia and the development of novel epididymal cell lines from infertile men. Biology of Reproduction , 83(4), 584–596. https://doi.org/10.1095/biolreprod.110.084459
- Dufresne, J., & Cyr, D. G. (2007). Activation of an SP binding site is crucial for the expression of claudin 1 in rat epididymal principal cells. Biology of Reproduction , 76(5), 825–832. https://doi.org/10.1095/biolreprod.106.057430
- Dufresne, J., Gregory, M., Pinel, L., & Cyr, D. G. (2022). Differential gene expression and hallmarks of stemness in epithelial cells of the developing rat epididymis. Cell and Tissue Research , 389(2), 327–349. https://doi.org/10.1007/s00441-022-03634-9
- Dufresne, J., St-Pierre, N., Viger, R. S., Hermo, L., & Cyr, D. G. (2005). Characterization of a novel rat epididymal cell line to study epididymal function. Endocrinology , 146(11), 4710–4720. https://doi.org/10.1210/en.2004-1634
- Franca, L. R., Auharek, S. A., Hess, R. A., Dufour, J. M., & Hinton, B. T. (2012). Blood-tissue barriers: Morphofunctional and immunological aspects of the blood-testis and blood-epididymal barriers. Advances in Experimental Medicine and Biology , 763, 237–259. https://doi.org/10.1007/978-1-4614-4711-5_12
- Gregory, M., & Cyr, D. G. (2019). Effects of prostaglandin E2 on gap junction protein alpha 1 in the rat epididymis. Biology of Reproduction , 100(1), 123–132. https://doi.org/10.1093/biolre/ioy171
- Gregory, M., Dufresne, J., Hermo, L., & Cyr, D. (2001). Claudin-1 is not restricted to tight junctions in the rat epididymis. Endocrinology , 142(2), 854–863. https://doi.org/10.1210/endo.142.2.7975
- Hermo, L., Barin, K., & Robaire, B. (1992). Structural differentiation of the epithelial cells of the testicular excurrent duct system of rats during postnatal development. The Anatomical Record , 233(2), 205–228. https://doi.org/10.1002/ar.1092330205
- Huch, M., Boj, S. F., & Clevers, H. (2013). Lgr5(+) liver stem cells, hepatic organoids and regenerative medicine. Regenerative Medicine , 8(4), 385–387. https://doi.org/10.2217/rme.13.39
- Jelinsky, S. A., Turner, T. T., Bang, H. J., Finger, J. N., Solarz, M. K., Wilson, E., Brown, E. L., Kopf, G. S., & Johnston, D. S. (2007). The rat epididymal transcriptome: Comparison of segmental gene expression in the rat and mouse epididymides. Biology of Reproduction , 76(4), 561–570. https://doi.org/10.1095/biolreprod.106.057323
- Jones, S. R., & Cyr, D. G. (2011). Regulation and characterization of the ATP-binding cassette transporter-B1 in the epididymis and epididymal spermatozoa of the rat. Toxicological Sciences , 119(2), 369–379. https://doi.org/10.1093/toxsci/kfq318
- Kim, J. K., Vinarsky, V., Wain, J., Zhao, R., Jung, K., Choi, J., Lam, A., Pardo-Saganta, A., Breton, S., Rajagopal, J., & Yun, S. H. (2012). In vivo imaging of tracheal epithelial cells in mice during airway regeneration. American Journal of Respiratory Cell and Molecular Biology , 47(6), 864–868. https://doi.org/10.1165/rcmb.2012-0164OC
- Kristensen, D. M., Nielsen, J. E., Kalisz, M., Dalgaard, M. D., Audouze, K., Larsen, M. E., Jacobsen, G. K., Horn, T., Brunak, S., Skakkebaek, N. E., & Leffers, H. (2010). OCT4 and downstream factors are expressed in human somatic urogenital epithelia and in culture of epididymal spheres. Molecular Human Reproduction , 16(11), 835–845. https://doi.org/10.1093/molehr/gaq008
- Leir, S. H., Yin, S., Kerschner, J. L., Xia, S., Ahmadi, S., Bear, C., & Harris, A. (2020). An organoid model to assay the role of CFTR in the human epididymis epithelium. Cell and Tissue Research , 381(2), 327–336. https://doi.org/10.1007/s00441-020-03208-7
- Li, L., & Clevers, H. (2010). Coexistence of quiescent and active adult stem cells in mammals. Science , 327(5965), 542–545. https://doi.org/10.1126/science.1180794
- Mandon, M., & Cyr, D. G. (2015). Tricellulin and its role in the epididymal epithelium of the rat. Biology of Reproduction , 92(3), 66. https://doi.org/10.1095/biolreprod.114.120824
- Mandon, M., Hermo, L., & Cyr, D. G. (2015). Isolated rat epididymal basal cells share common properties with adult stem cells. Biology of Reproduction , 93(5), 115. https://doi.org/10.1095/biolreprod.115.133967
- Martan, J., Risley, P. L., & Hruban, Z. (1964). Holocrine cells of the human epididymis. Fertility and Sterility , 15, 180–187. https://doi.org/10.1016/s0015-0282(16)35166-4
- Mital, P., Hinton, B. T., & Dufour, J. M. (2011). The blood-testis and blood-epididymis barriers are more than just their tight junctions. Biology of Reproduction , 84(5), 851–858. https://doi.org/10.1095/biolreprod.110.087452
- Mou, H., Vinarsky, V., Tata, P. R., Brazauskas, K., Choi, S. H., Crooke, A. K., Zhang, B., Solomon, G. M., Turner, B., Bihler, H., Harrington, J., Lapey, A., Channick, C., Keyes, C., Freund, A., Artandi, S., Mense, M., Rowe, S., Engelhardt, J. F., … Rajagopal, J. (2016). Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell , 19(2), 217–231. https://doi.org/10.1016/j.stem.2016.05.012
- Orgebin-Crist, M. C. (1968). Maturation of spermatozoa in the rabbit epididymis: Delayed fertilization in does inseminated with epididymal spermatozoa. Journal of Reproduction & Infertility, 16(1), 29–33. https://doi.org/10.1530/jrf.0.0160029
- Papp, S., Robaire, B., & Hermo, L. (1995). Immunocytochemical localization of the Ya, Yc, Yb1, and Yb2 subunits of glutathione S-transferases in the testis and epididymis of adult rats. Microscopy Research and Technique , 30(1), 1–23. https://doi.org/10.1002/jemt.1070300102
- Pinel, L., & Cyr, D. G. (2021). Self-renewal and differentiation of rat epididymal basal cells using a novel in vitro organoid model. Biology of Reproduction , 105(4), 987–1001. https://doi.org/10.1093/biolre/ioab113
- Pinel, L., Mandon, M., & Cyr, D. G. (2019). Tissue regeneration and the epididymal stem cell. Andrology , 7(5), 618–630. https://doi.org/10.1111/andr.12635
- Robaire, B., & Hermo, L. (1988). Efferent ducts, epididymis and vas deferens: Structure, functions and their regulation In E. Knobil & J. Neill (Eds.), The physiology of reproduction. (pp. 999–1080). Raven Press.
- Robaire, B., Hinton, B. T., & Orgebin-Crist, M. C. (2015). The epididymis. In T. Plant & A. Zeleznik (Eds.), Knobil and Neill's reproductive physiology (pp. 691–771). Academic Press.
- Sangiorgi, E., & Capecchi, M. R. (2008). Bmi1 is expressed in vivo in intestinal stem cells. Nature Genetics , 40(7), 915–920. https://doi.org/10.1038/ng.165
- Sharma, U., Conine, C. C., Shea, J. M., Boskovic, A., Derr, A. G., Bing, X. Y., Belleannee, C., Kucukural, A., Serra, R. W., Sun, F., Song, L., Carone, B. R., Ricci, E. P., Li, X. Z., Fauquier, L., Moore, M. J., Sullivan, R., Mello, C. C., Garber, M., & Rando, O. J. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science , 351(6271), 391–396. https://doi.org/10.1126/science.aad6780
- Shum, W. W., Da, S. N., McKee, M., Smith, P. J., Brown, D., & Breton, S. (2008). Transepithelial projections from basal cells are luminal sensors in pseudostratified epithelia. Cell , 135(6), 1108–1117. https://doi.org/10.1016/j.cell.2008.10.020
- Shum, W. W., Hill, E., Brown, D., & Breton, S. (2013). Plasticity of basal cells during postnatal development in the rat epididymis. Reproduction , 146(5), 455–469. https://doi.org/10.1530/REP-12-0510
- Shum, W. W., Ruan, Y. C., Da, S. N., & Breton, S. (2011). Establishment of cell-cell cross talk in the epididymis: Control of luminal acidification. Journal of Andrology , 32(6), 576–586. https://doi.org/10.2164/jandrol.111.01297
- Sipila, P., Shariatmadari, R., Huhtaniemi, I. T., & Poutanen, M. (2004). Immortalization of epididymal epithelium in transgenic mice expressing simian virus 40 T antigen: Characterization of cell lines and regulation of the polyoma enhancer activator 3. Endocrinology , 145(1), 437–446. https://doi.org/10.1210/en.2003-0831
- Sullivan, R. (2015). Epididymosomes: A heterogeneous population of microvesicles with multiple functions in sperm maturation and storage. Asian Journal of Andrology , 17(5), 726–729. https://doi.org/10.4103/1008-682X.155255
- Sullivan, R., Legare, C., Lamontagne-Proulx, J., Breton, S., & Soulet, D. (2019). Revisiting structure/functions of the human epididymis. Andrology , 7(5), 748–757. https://doi.org/10.1111/andr.12633
- Sun, E. L., & Flickinger, C. J. (1980). Morphological characteristics of cells with apical nuclei in the initial segment of the adult rat epididymis. The Anatomical Record , 196(3), 285–293. https://doi.org/10.1002/ar.1091960304
- Turner, T. T. (1995). On the epididymis and its role in the development of the fertile ejaculate. Journal of Andrology , 16(4), 292–298. https://doi.org/10.1002/j.1939-4640.1995.tb00532.x
- Turner, T. T. (2002). Necessity's potion: Inorganic ions and small organic molecules in the epididymal lumen. In B. H. Robaire & B. T. Hinton (Ed.), The epididymis: From molecules to clinical practice (pp. 131–150). Plenum Press.
- Turner, T. T. (2008). De Graaf's thread: The human epididymis. Journal of Andrology , 29(3), 237–250. https://doi.org/10.2164/jandrol.107.004119
- Turner, T. T., Bomgardner, D., Jacobs, J. P., & Nguyen, Q. A. (2003). Association of segmentation of the epididymal interstitium with segmented tubule function in rats and mice. Reproduction , 125(6), 871–878. https://doi.org/10.1530/rep.0.1250871
- Veri, J. P., Hermo, L., & Robaire, B. (1993). Immunocytochemical localization of the Yf subunit of glutathione S-transferase P shows regional variation in the staining of epithelial cells of the testis, efferent ducts, and epididymis of the male rat. Journal of Andrology , 14(1), 23–44. https://doi.org/10.1002/j.1939-4640.1993.tb00366.x
- Voisin, A., Damon-Soubeyrand, C., Bravard, S., Saez, F., Drevet, J. R., & Guiton, R. (2020). Differential expression and localisation of TGF-beta isoforms and receptors in the murine epididymis. Scientific Reports , 10(1), 995. https://doi.org/10.1038/s41598-020-57839-5
- Yan, K. S., Chia, L. A., Li, X., Ootani, A., Su, J., Lee, J. Y., Su, N., Luo, Y., Heilshorn, S. C., Amieva, M. R., Sangiorgi, E., Capecchi, M. R., & Kuo, C. J. (2012). The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proceedings of the National Academy of Sciences of the United States of America , 109(2), 466–471. https://doi.org/10.1073/pnas.1118857109
Citing Literature
Number of times cited according to CrossRef: 2
- Junyu Nie, Hao Chen, Xiuling Zhao, Advancement and Potential Applications of Epididymal Organoids, Biomolecules, 10.3390/biom14081026, 14 , 8, (1026), (2024).
- Mariana Weigel Muñoz, Débora J. Cohen, Vanina G. Da Ros, Soledad N. González, Abril Rebagliati Cid, Valeria Sulzyk, Patricia S. Cuasnicu, Physiological and pathological aspects of epididymal sperm maturation, Molecular Aspects of Medicine, 10.1016/j.mam.2024.101321, 100 , (101321), (2024).

