Targeted ExSeq -- Sequencing Library Preparation
Yi Cui, Anubhav Sinha, Ed Boyden, Asmamaw T. Wassie, Fei Chen
expansion microscopy
in situ sequencing
expansion sequencing
targeted ExSeq
ExSeq
spatial transcriptomics
spatial omics
spatially resolved transcriptomics
Disclaimer
This protocol is shared under the HTAN Internal Data and Materials Sharing Agreement and is provided as is. See section 14 of full agreement for full details.
This protocol is shared as an Open Access protocol (under HTAN's definitions). This protocol is Subject to IP Restrictions.
Abstract

This protocol accompanies Expansion Sequencing (ExSeq), describing the process of targeted ExSeq library preparation for a sample that has been processed according to a Targeted ExSeq Tissue Preparation protocol. The steps described here are a generalization of the protocol used in figures 4-6 of the paper, and represent our recommendations for future users of the technology.
The flowchart in Fig. 1A depicts the library preparation workflow. Fig. 1B is a summary of the product, in which padlock probes are amplified to form amplicon concatamers. The net result of the process is that barcode sequences are delivered to transcripts of interest and locally amplified hundreds to thousands of times.
The process of library preparation encompasses the following steps. (1) Oligonucleotide padlock probes bearing barcode sequences hybridize to RNA transcripts of interest (Step 8). (2) SplintR Ligase, which can has RNA-splinted DNA ligase activity, ligates adjacent ends of padlock probes, forming circular DNA molecules (Step 9). (3) A universal primer hybridizes to all padlock probes (Step 10). (4) Rolling Circle Amplification initiates from the primers, and repeatedly copies the sequence of the padlock probes, forming an amplicon (Step 11). (5) BS(PEG)9 covalently cross-links the amplicon to itself, stabilizing the amplicon (Steps 12-13) during in situ sequencing. At this point, the sample is now ready for downstream detection (i.e. via hybridizing fluorophore-labeled oligos to the amplicon (Step 14), and in situ sequencing).
This protocol was used (with no modifications) to profile human metastatic breast cancer biopsies as a part of the Human Tumor Atlas Pilot Project (HTAPP).
Before start
Prepare stock solutions/reagents as described in the section Preparation of Stock Solutions (Steps 1-3), and Preparation of Oligonucleotide Solutions (Steps 4-6).
The assumed input to the protocol is a sample processed by by a Targeted ExSeq Tissue Preparation, as described in Step 7.
Steps
Preparation of Stock Solutions
Common Stock Solutions
-
1X PBS
-
2X SSC (Saline-Sodium Citrate) buffer
-
Wash buffer: 2X SSC, 20% formamide (v/v) in water
-
Wash-10 buffer: 2X SSC, 10% formamide (v/v) in water
-
Stripping solution: 80% formamide (v/v), 0.01% Triton X-100 (v/v) in water
Suggested volumes to prepare: >10 mL. All buffers can be stored at Room temperature. Wash buffer should be used within one month of preparation.
4 mM aminoallyl dUTP (aadUTP)
Prepare a 100 μL aliquot of 4 mM aminoallyl dUTP and store at -20°C.
250 mM BS(PEG)9
Upon receipt of BS(PEG)9, store in -20°C freezer in original packing.
To aliquot for use: allow BS(PEG)9 vial to warm up to room temperature completely to avoid condensation. Add 465 µL anhydrous DMSO, vortex, and aliquot ~20 µL volumes into PCR tubes.
Store PCR tubes at -20°C in a Parafilm-sealed 50 mL tube containing Drierite.
BS(PEG)9 aliquots should be used within one month from preparation.
Preparation of Oligonucleotide Solutions
Pooled Padlock Probes
A more complete discussion of the process of pooling padlock probes is in the probe generation protocol. Briefly, padlock probes should be pooled together such that the concentration of each individual padlock probe is the same, and that the total probe concentration is maximized, typically 200 µM.
We use the per-probe concentration as a convenient metric when calculating dilutions.
As an example, if 40 genes are being interrogated with 16 padlocks/gene, the probes should be pooled together to have a total concentration of 200 µM, with the concentration per probe being 200 µM/(40 genes * 16 probes/gene) = 312.5 nM per probe.
100 µM Rolling Circle Amplification (RCA) Primer
The sequence of the RCA primer is TCT TCA GCG TTC CCG AGA, where * denotes phosphorothioate backbone modification. This modification can be included by all major DNA synthesis companies.
Resuspend lyophilized DNA in water to prepare a 100 µM stock solution. Prepare a ~100 µL working stock solution aliquot.
Store RCA primer at -20°C.
100 µM Amplicon Detection Oligonucleotide Probe
The sequence of the amplicon detection oligo is /5Cy3/TCTCGGGAACGCTGAAGA, where /5Cy3/ denotes a 5' Cy3 dye. This dye can be replaced with Alexa 546, or any other dye that works well for your imaging setup. Due to the dye-modification, this oligo typically requires HPLC purification.
Resuspend lyophilized DNA in water to prepare a 100 µM stock solution. Prepare a ~25 µL working stock solution aliquot.
Store amplicon detection oligo at -20°C.
Preparation of Samples
Initial Preparation of Samples
We assume that samples have been prepared as described by an appropriate Targeted ExSeq Tissue Preparation protocol. In particular, we assume that the total sample thickness after re-embedding and passivation is <350 µm. Thicker samples will require additional optimization.
We assume that the gels have been trimmed and cut to the appropriate size, and are currently stored in 1X PBS in PCR tubes or in microcentrifuge tubes.
See note in Guidelines on volumes. Briefly, volumes described below are 200 µL unless noted otherwise, appropriate for samples in PCR tubes. For all non-wash steps for samples in PCR tubes, volumes can be scaled down to 50-100 µL, as long as the gel is fully submerged. Washes in PCR tubes should remain at 200 µL. For samples in microcentrifuge tubes, recipes can be scaled up appropriately to completely submerge the gel, typically 400-600 µL. Washes in microcentrifuge tubes should be 500-1000 µL in volume.
Since multiple samples are typically processed in parallel, we use the term gels in the following steps. Note that each gel should be processed in its own independent tube.
Padlock Probe Hybridization
Padlock Probe Hybridization
The first step of library preparation is to hybridize padlock probes to the RNA transcripts of interest within the expanded samples.
Pre-hybridization
Pre-hybridize gels by washing with wash buffer for 0h 30m 0s at Room temperature.
Preparation of Hybridization Buffer
During the pre-hybridization step, prepare the hybridization buffer. The hybridization buffer consists of the oligonucleotide padlock probes in 2X SSC, 20% formamide.
The concentration of the pooled padlock probes can be variable. We have observed increasing yield with increased per-probe concentration. However, in experiments with a very large number of probes, the per-probe concentration will remain low, due to the total number of probes in solutions. We suggest 100 nM per probe as a starting point for optimization.
Hybridization Mix:
| A | B | C | D |
|---|---|---|---|
| Solution | Stock Concentration | Volume | Final Concentration |
| SSC buffer | 20X | 20 µL | 2X |
| Formamide | 100% | 40 µL | 20% |
| Pooled Padlock Probes | X nM per probe | x µL | variable; suggested 100 nM per probe |
| Water | (140 - x) µL | ||
| Total | 200 µL |
Padlock Probe Hybridization
Incubate gels with hybridization mix for at 37°C.
Wash Buffer Washes
Wash gels with wash buffer for 0h 30m 0s at 37°C.
For concentrated probe libraries (>100 uM final total probe concentration), this can be increased to 4-5 washes.
PBS Wash
Wash gels with 1X PBS for 0h 30m 0s at 37°C.
Proceed directly to Padlock Probe Ligation.
Padlock Probe Ligation
Enzymatic Ligation of Padlock Probes
In this step of the protocol, the hybridized padlock probes are enzymatically ligated by SplintR Ligase, forming circular DNA molecules that can subsequently be amplified by rolling circle amplification. After ligation, the padlock probes are topologically intertwined with the RNA transcripts, as the homology region is 32 bp in length.
Of particular importance here is the pre-ligation step, in which SplintR ligase diffuses into the sample at 4°C with minimal ligation activity. We found that both extended pre-ligation and pre-amplification steps with SplintR ligase and Phi29 (respectively) are needed to generate amplicons uniformly throughout the volume of the tissue, without any Z-dependence.
Buffer Pre-equilibration
Wash the gels with 1X SplintR Ligase Buffer for 0h 30m 0s at Room temperature.
Pre-ligation
Prepare ligation mix at 4°C (on a cold block or on ice).
Ligation mix:
| A | B | C | D |
|---|---|---|---|
| Reagent | Stock Concentration | Volume | Final Concentration |
| Water | 170 µL | ||
| SplintR Ligase Buffer | 10X | 20 µL | 1X |
| SplintR Ligase | 25 U/µL | 10 µL | 1.25 U/µL |
| Total | 200 µL |
Incubate gels with pre-ligation mix for 6h 0m 0s at 4°C.
Note: in our testing, the duration of the pre-ligation step may potentially be shortened to three hours, or extended to overnight without adverse effects. We recommend six hours as a starting point for further optimization.
--- Possible Pause Point ---
The pre-ligation step can be run overnight.
Enzymatic Ligation
Prepare a fresh volume of ligation mix (described in previous substep). Volume can be scaled as described above.
Incubate gels with ligation mix for 6h 0m 0s at 37°C.
Note: in our testing, ligation can be performed overnight if necessary. We recommend six hours, to minimize RNA degradation.
--- Possible Pause Point ---
The ligation step can be run overnight.
Ligation Termination
Wash the gels with 2X SSC buffer for 0h 30m 0s at Room temperature.
Proceed directly to RCA Primer Hybridization.
RCA Primer Hybridization
RCA Primer Hybridization
In this step, we hybridize a universal primer for rolling circle amplification to the constant sequence of the backbone of the padlock probe. The RCA primer has phosphorothioate modifications to prevent degradation by the strong 3'->5' exonuclease activity of Phi29.
Pre-hybridization
Pre-hybridize gels by washing with wash buffer for 0h 15m 0s at 37Room temperature.
RCA Primer Hybridization
Prepare the RCA primer hybridization mix by diluting the 100 µM RCA primer stock by 1:200 in wash buffer, forming a 500 nM RCA primer solution.
Incubate gels in RCA primer hybridization mix for 2h 0m 0s at 37°C.
Note: in our testing, RCA primer hybridization can be run overnight if necessary. We recommend two hours, to minimize RNA degradation.
--- Possible Pause Point ---
The RCA primer hybridization can be run overnight.
Wash Buffer Wash
Wash gels with wash buffer for 0h 30m 0s at 37°C.
PBS Wash
Wash gels with 1X PBS for 0h 15m 0s at 37°C.
Proceed directly to Rolling Circle Amplification (RCA).
Rolling Circle Amplification (RCA)
Rolling Circle Amplification
In this step, the ligated padlock probes are amplified using rolling circle amplification (RCA), forming amplicons (colloquially called "RCA colonies" or "rolonies").
Similar to the ligation step, we have an extended pre-amplification step, in which the Phi29 enzyme diffuses into the sample at 4°C. During RCA, aminoallyl dUTP is incorporated into the amplicons, enabling subsequent crosslinking by BS(PEG)9.
Buffer Pre-equilibration
Wash the gels with 1X Phi29 Buffer for 0h 15m 0s at Room temperature.
Pre-amplification
Prepare pre-amplification mix at 4°C (on a cold block or on ice).
Pre-amplification mix:
| A | B | C | D |
|---|---|---|---|
| Reagent | Stock Concentration | Volume | Final Concentration |
| Water | 178 µL | ||
| Phi29 Buffer | 10X | 20 µL | 1X |
| Phi29 DNA Polymerase | 100 U/µL | 2 µL | 1 U/µL |
| Total | 200 µL |
Incubate gels with pre-amplification mix for 6h 0m 0s at 4°C.
Note: in our testing, the duration of the pre-amplification step may potentially be shortened to three hours, or extended to overnight without adverse effects. We recommend six hours as a starting point for further optimization as we have used this duration most often.
--- Possible Pause Point ---
The pre-amplification step can be run overnight.
Rolling Circle Amplification
Prepare RCA mix at 4°C (on a cold block or on ice).
RCA mix:
| A | B | C | D |
|---|---|---|---|
| Reagent | Stock Concentration | Volume | Final Concentration |
| Water | 171 µL | ||
| Phi29 Buffer | 10X | 20 µL | 1X |
| dNTP | 10 mM each nucleotide | 5 µL | 250 µM |
| Aminoallyl dUTP (aadUTP) | 4 mM | 2 µL | 40 µM |
| Phi29 DNA Polymerase | 100 U/µL | 2 µL | 1 U/µL |
| Total | 200 µL |
Incubate gels with RCA mix for 6h 0m 0s at 30°C.
--- Pause Point ---
The RCA reaction runs overnight.
PBS Wash
Terminate the RCA by washing the sample with 1X PBS for 0h 30m 0s at Room temperature.
Proceed directly to BS(PEG)9 Cross-Linking
BS(PEG)9 Cross-Linking
BS(PEG)9 Cross-Linking
To prevent amplicon movement across multiple rounds of in situ sequencing, amplicons are covalently anchored to themselves, resulting in a cross-linked DNA amplicon that is enmeshed with the expansion gel.
To accomplish this, aminoallyl dUTP (aadUTP) is included in the RCA mix. In this step, the NHS esters of BS(PEG)9 react with the primary amines of the allylamine groups, crosslinking the amplicon to itself. The structures of aadUTP and BS(PEG)9 are shown below.
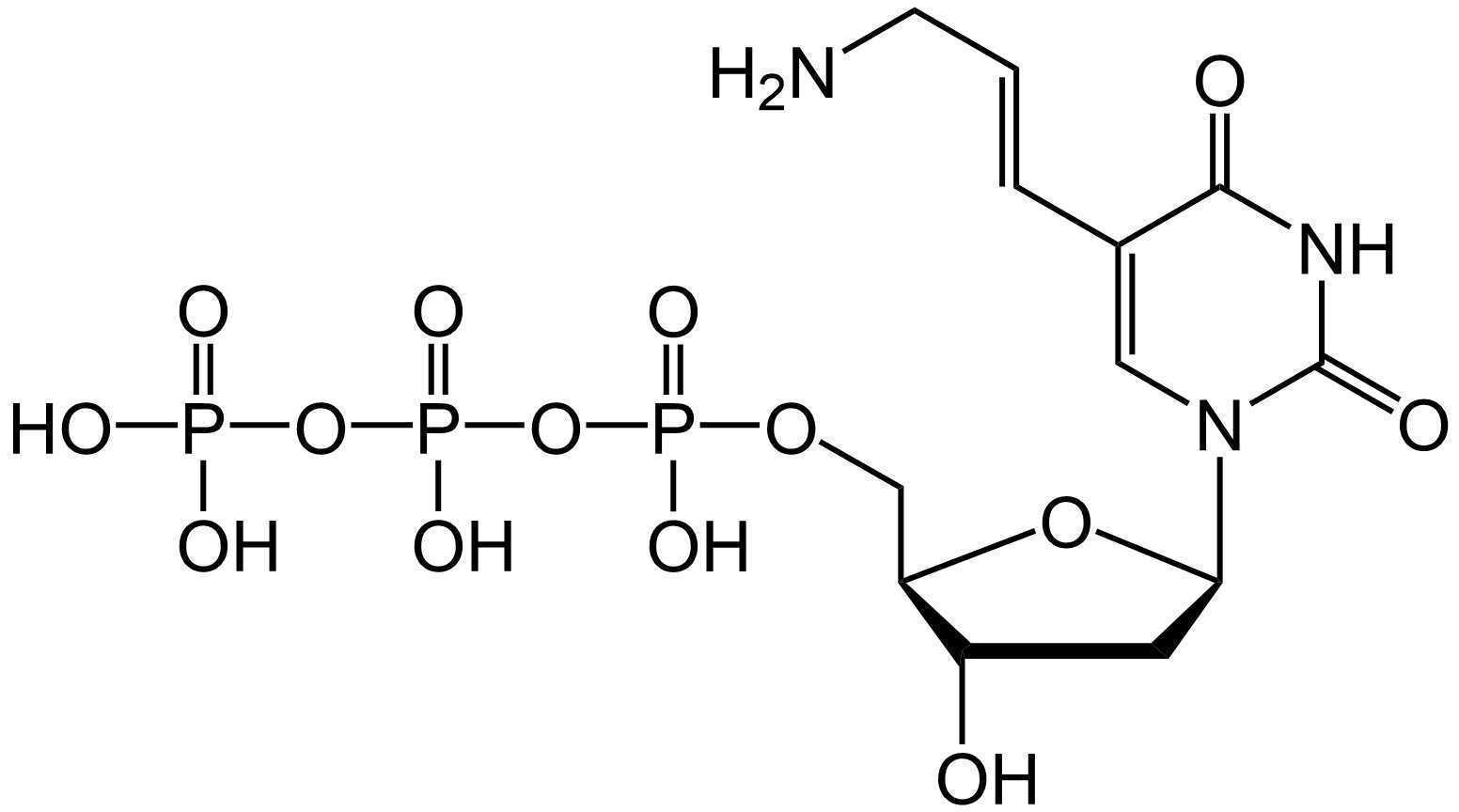
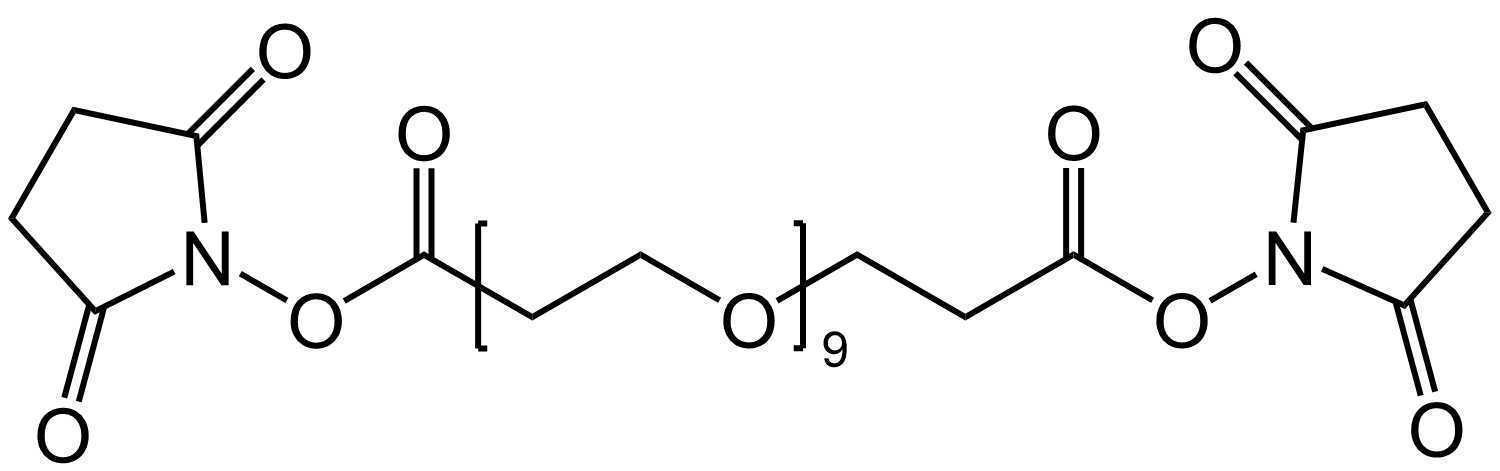
Amplicon Cross-linking
Thaw one aliquot of reconstituted 250 mM BS(PEG)9 at room temperature.
Prepare crosslinking mix:
| A | B | C | D |
|---|---|---|---|
| Reagent | Stock Concentration | Volume | Final Concentration |
| PBS | 1X | 196 µL | ~1X |
| BS(PEG)9 | 250 mM | 4 µL | 5 mM |
| Total | 200 µL |
Incubate gels with crosslinking mix for 2h 0m 0s at Room temperature.
Tris Wash
Quench reaction by washing gels with 1 M Tris pH 8 for 0h 15m 0s at Room temperature.
PBS Wash
Wash gels with 1X PBS for 0h 30m 0s at Room temperature.
[OPTIONAL] Long Term Storage
At this point, the library preparation process is complete, and the amplicons are stable.
For long-term storage, replace the 1X PBS solution once more, then store gels in well-sealed containers at 4°C for up to several months.
Universal Amplicon Detection Hybridization
Amplicon Detection Hybridization
A rapid quality control assay for the library preparation process is to visualize all amplicons within the specimen by hybridizing a fluorophore-labeled oligonucleotide probe to the amplicon constant region, thereby labeling all amplicons for imaging.
Pre-hybridization
Wash gels with Wash-10 buffer for 0h 20m 0s at Room temperature.
Detection Probe Hybridization
Prepare the amplicon detection probe hybridization mix by diluting the 100 µM amplicon detection probe stock by 1:1000 in Wash-10 buffer, forming a 100 nM amplicon detection probe solution.
Incubate gels in amplicon detection probe solution for 1h 0m 0s at 37°C.
Wash-10 Wash
Wash gels with Wash-10 buffer for 0h 20m 0s at 37°C.
PBS Wash
Wash gels with 1X PBS for 0h 20m 0s at 37°C.
DAPI Staining
Stain gels with DAPI in 1X PBS. Recipe for 1 mL DAPI solution:
| A | B | C | D |
|---|---|---|---|
| Reagent | Stock Concentration | Volume | Final Concentration |
| PBS | 1X | 999 μL | 1X |
| DAPI | 1 mg/mL | 1 μL | 1 mg/L |
| Total | 1000 μL |
Stain gel with DAPI solution for 0h 15m 0s at Room temperature.
Wash with 1X PBS for 0h 10m 0s at Room temperature.
Remove 1X PBS thoroughly to prevent gel movement during imaging.
Imaging
After PBS wash, remove excess PBS and use a clean paintbrush to transfer gel to a glass-bottom 6-well or 24-well plate for imaging.
Image as is appropriate for the dye used to visualize amplicons. Individual amplicons should be visible at 10X and higher magnification air-objectives, with significantly higher image quality with 20X and higher magnification, and high NA, water/oil immersion objectives. Our standard objective for expansion microscopy is the Nikon 40X Long Working Distance, Water Immersion, NA 1.15 objective (CFI ApoLWD Lambda S 40XC WI).
Stripping Detection Oligo
Transfer the gel from the imaging plate to an appropriate smaller container, i.e. PCR tube or microcentrifuge tube.
Heat an appropriate amount of stripping solution to 80°C. The stripping solution can be heated in a thermocycler. The total volume pre-heated should be enough for three full washes, i.e. 600 μL for gels in PCR tubes.
Wash the sample with pre-heated stripping solution for 0h 10m 0s. For each wash, use pre-heated stripping solution, but keep the sample at Room temperature .
PBS Washes and Storage
Wash gels with 1X PBS for 0h 10m 0s at Room temperature. Then, store as described in Step 13.

