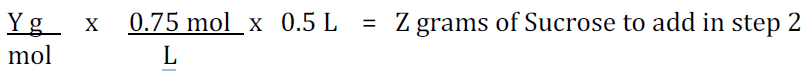Sucrose lysis buffer
Colleen Kellogg
Disclaimer
Abstract
This protocol describes the preparation of sucrose lysis buffer to preserve DNA on sterivex filters. As part of the Hakai Institute Ocean Observing Program, biomolecular samples have been collected weekly, from 0 m to near bottom (260 m), to genetically characterize plankton communities in the Northern Salish Sea since 2015. This protocol is developed to work across all domains of life, from viruses to prokaryotes to eukaryotes, allowing for both amplicon sequencing and shotgun sequencing. The protocol is part of the Hakai Institute's pipeline to analyze microbial and environmental DNA from seawater samples and is implemented as a standard procedure for ongoing sampling programs.
Before start
Steps
Preparations
You will need:
- MilliQ Water
-500 mL bottle top filtration unit
Final concentrations of chemicals in SLB:
EDTA: 40 mM
Tris: 50 mM
Sucrose: 0.75 M
Calculations
Calculation of Tris or EDTA:
Use the equation C1V1 = C2V2
Eg for EDTA:
(0.5M)(X mL) = (0.04M)(500 mL)
Solve for X
((0.04M)(500 mL)/(0.5M)) = 40 mL of 0.5M EDTA
Methods
Add the appropriate amount of Sucrose calculated above and place it in a clean bottle or beaker.
Add 40 mL of 0.5M EDTA to the beaker.
Add 25mL of 1M Tris to the beaker.
Add milliQ water to about the 400 mL line.
Add a stir bar and dissolve all the powder.
Top up water to 500 mL. (no need to pH this one!)
Filter-sterilize and label bottle.