Single Cell Isolation of Human Lung Organoids
Morris Baumgardt, Stefan Hippenstiel, Andreas C. Hocke, Katja Hönzke, Maren Hülsemann, Anna Löwa, Benedikt Obermayer, Emanuel Wyler
single cell isolation
human lung organoids
single cell RNA sequencing
methanol fixation
rehydration of methanol fixed cells
Disclaimer
Informed written consent was obtained from all volunteers and the study was approved by the Charité Ethics Committee (project 451, EA2/079/13).
Abstract
This protocol describes the single cell isolation of infected human alveolar-like organoids in order to perform single cell RNA sequencing or flow cytometry. The protocol focussed on the detailed preparation of samples to prepare single cells for further processing according to the protocols from 10x Genomics for single cell sequencing. The described steps also include the optional fixation of single cells with methanol. In that case, the protocol describes how to rehydrate single cell after methanol fixation to process them for single cell sequencing.
Before start
Steps
Single cell dissociation
Remove organoid medium from two wells and add 1mL cold base medium per well (300,000 cells per well is a good amount to have sufficient cells even after the dissociation process.
Transfer organoids with a 1000 µL pipette to a 2 mL reagent tube and spin down at 300x g,4°C.
Very carefully discard the supernatant with a 1000 µL pipette.
Resuspend the cell pellet in 1mL TrypLE Express/well (initally combined).
Incubate at 37°C for 0h 15m 0s carefully vortex every 5 mins, stop the reaction with 1mL of base medium.
Spin down at 300x g,4°C.
Very carefully discard the supernatant.
Resuspend the cell pellet in 1mL cold base medium.
Resuspend the whole supernatant 3 times with 27G needle and 1 mL syringe.
Filter cells through the cap of FACS tubes (40 µm pore size).
Count cells with the use of Trypan Blue staining. Make sure a single cell solution is provided. According to 10x Genomics recommendation a high-quality cell suspension has high cell viability (>90% ideally, >70% is acceptable), low cell debris and low cell clumping.
Proceed with methanol fixation or prepare samples immediately according to 10x Genomics protocol for RNA-Sequencing or flow cytometry.
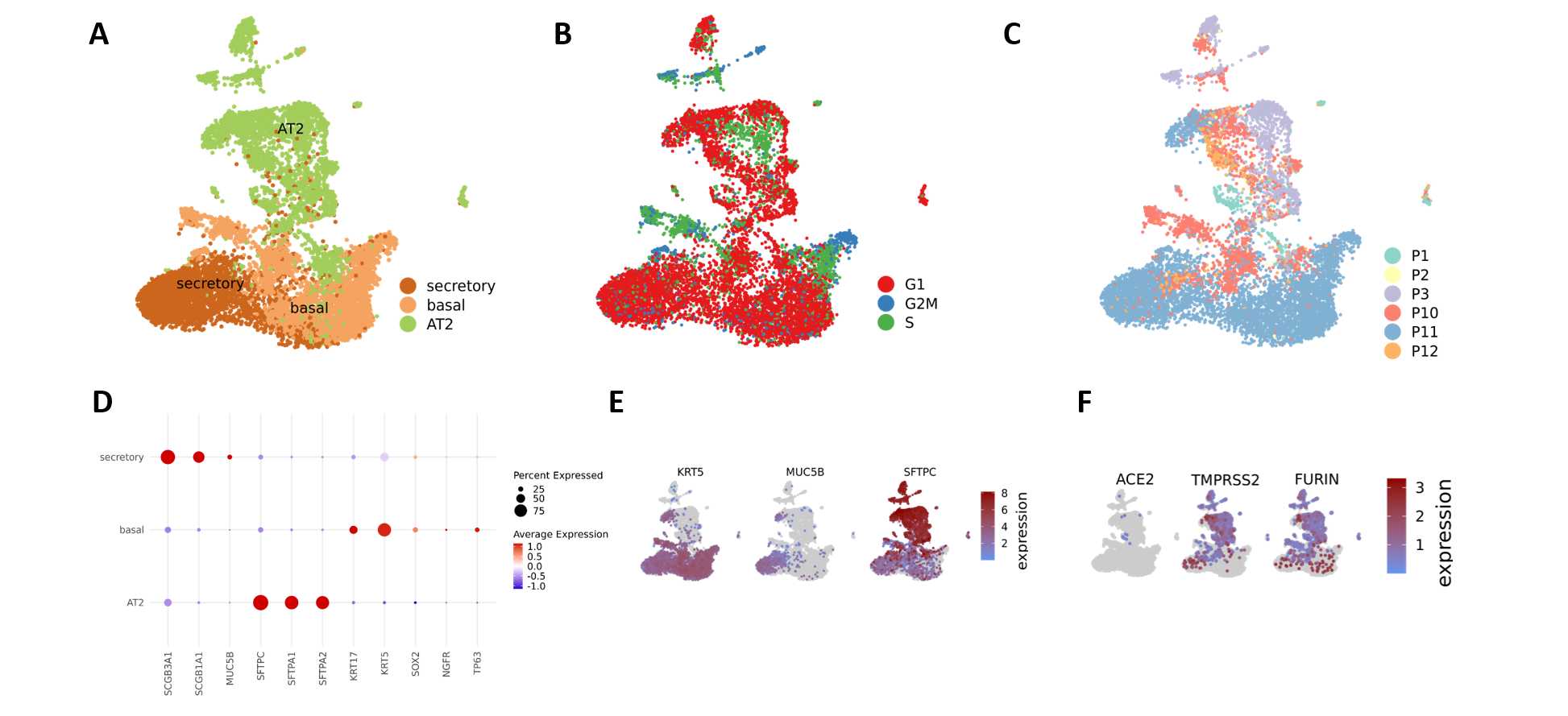
The Cell Ranger Software Suite (Version 3.1.0) was used to process raw sequencing data with the GRCh38 reference for the control samples, and GRCh38 augmented by the SARS-CoV-2 (NC_045512.2). Viral transcripts including 3'UTR sequences were extracted from the Genbank records and added to the Gencode v33 reference. We used CellBender to remove background RNA and scrublet to identify doublets. Single-cell RNA sequencing data analysis was performed in R (version 3.6.3) with Seurat (version 3.2.1). Cells with at least 500 and less than 5000 detected genes and less than 10% mitochondrial content were combined from each library and library depth (total number of UMIs) was regressed out when scaling data. We additionally regressed out cell cycle scores calculated with Seurat's CellCycleScoring function. After automated clustering, DoubletFinder was used to again identify likely cell doublets, and cluster annotation was performed with Seurat's ‘TransferData’ workflow using the Human Lung Cell Atlas reference dataset, using only epithelial cells for the organoid data.
Methanol fixation (optional)
In case of scRNA-sequencing: Ideally methanol fixation should be avoided and the samples should be immediately processed for library preparation according to the 10x Genomics protocol (Chromium Next GEM Single Cell 3’ Reagent Kits v3.1 (Dual Index) with Feature Barcode technology for Cell Multiplexing User Guide).
Centrifuge the dissociated cells at 300x g,4°C.
Remove the supernatant without disrupting the cell pellet.
Add 1mL 1x PBS and gently resuspend pellet 10 times with 1000 µL pipette tip or until cells are resuspended. Each tube should have 1-2 x106 cells (1 well of organoids usually contains up to 0.5 x106cells).
Repeat washing steps 14, 15 and 16.
Centrifuge at 300x g,4°C.
Remove the supernatant without disrupting the cell pellet.
Add 200µL chilled 1x PBS/1x106 cells and gently resuspend pellet 10x with 1000 µL pipette tip or until cells are resuspended.
Add 800µL chilled 100% methanol/1x106 cells. To avoid clumping of cells, add methanol drop by drop while gently stirring the cell suspension with the pipette tip in the tubes.
Incubate for 0h 30m 0s at -20°C.
Export from the BSL3 and store fixed cells either at -20°C or -80°C (stability has been shown for both termperatures for up to 6 weeks).
Rehydration of methanol-fixed cells
Place the microcentrifuge tube containing the methanol-fixed cells on ice to equilibrate to 4°C for 0h 5m 0s.
Centrifuge fixed cells at 1000x g,4°C.
Remove the supernatant without disrupting the cell pellet.
Based on starting cell concentration and assuming ~50% cell loss, add an appropriate volume of Wash-Resuspension Buffer to obtain a concentration of 700-1,200 cells/µL. Gently pipette mix using a regular-bore pipette tip until a single cell suspension is achieved.
Pass the sample through a 40 μm Flowmi Cell.
Count cells.
Proceed immediately with the 10x Genomics Single Cell protocols. Delay in proceeding may result in RNA loss.

