SeV standard and copyback genomes PCR Protocols
Carolina Lopez
Abstract
Protocols for the detection of Sendai Virus (SeV) standard and copy-back genomes by PCR
Before start
PRINCIPLES BEHIND THE PROCEDURE MUST BE UNDERSTOOD. PLEASE CONSULT WITH EXPERIENCED LAB MEMBER THE FIRST TIME YOU USE THIS PROCEDURE. UPDATE AS A GENERAL PROCEDURE AS NECESSARY BUT DO NOT MODIFY WITH SPECIFICS TO YOUR PROJECT, INSTEAD DOWNLOAD AND PASTE A MODIFIED COPY IN YOUR NOTEBOOK.
Steps
Reverse Transcription (RT)
*Different RT kits and steps for cbVG and Genome
**1/3) cbVG: Use SuperScript III Kit**
Procedure
*RT reaction can be completed in a 10µL reaction or a 20µLreaction. The 10µLreaction is preferred, if RNA concentrations allow, to help conserve reagents.
1. Start with `1µg` of RNA
`10µL` reaction: dilute in `4µL` dH<sub>2</sub>O
`20µL` reaction: dilute in `8µL` dH<sub>2</sub>O
2. Pre-mix `10micromolar (µM)` primer and dNTPs
`10µL` reaction: `0.5µL` of each, per sample
`20µL` reaction: `1µL`of each, per sample
3. Add Primer/dNTP mix to the RNA sample
`10µL` reaction: `1µL`
`20µL` reaction: `2µL`
4. Mix and spin down
5. Incubate at `65°C` for `0h 10m 0s`
Program in PCR Machine: RT-DI1
6. Prepare the mix (see TABLE below)
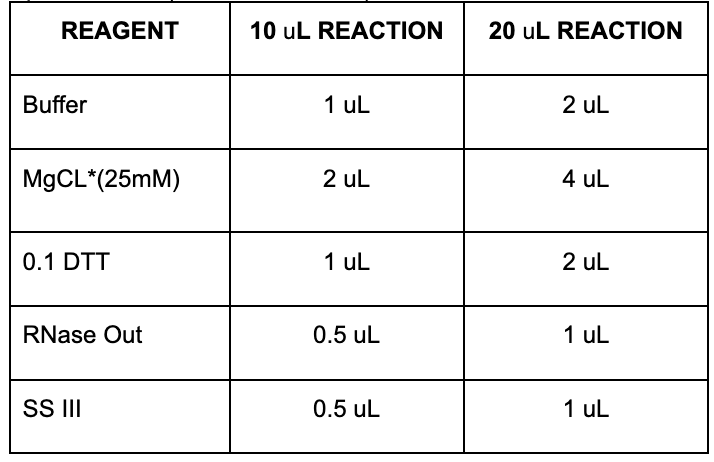
• Add the SIII enzyme separately to each sample, don’t include it in the mix.
7. Add mix to sample, vortex briefly, and spin down
`10µL` reaction: `5µL`
`20µL` reaction: `10µL`
8. Incubate at `50°C` for `0h 50m 0s` then `85°C` for `0h 5m 0s`
Program in PCR Machine: RT-DI2
9. After program is finished keep at `-20°C` or on ice for at least `0h 5m 0s`
————————————— Move to PCR Hood (for all later steps)
————————————— (Clean PCR hood with 10% bleach. Wipe down pipette with bleach in between samples.)
10. Add RNase H
`10µL` reaction: `0.5µL`
`20µL` reaction: `1µL`
11. Incubate at `37°C` for `0h 20m 0s`
Program in PCR Machine: RNaseH
12. Keep on ice for at least `0h 5m 0s` then you can do the PCR or store at `-20°C`
*Different RT kits and steps for cbVG and Genome 2/3) Genome: Use Roche Kit
Procedure
*RT reaction can be completed in a 10µL reaction or a 20µL reaction. The 10 ∝L reaction is preferred, if RNA concentrations allow, to help conserve reagents.
1. Start with `1000ng` of RNA diluted in dH<sub>2</sub>O
`10µL` reaction: dilute in `3µL` dH<sub>2</sub>O
`20µL` reaction: dilute in `6µL` dH<sub>2</sub>O
2. Add primer:
`10µL` reaction: `3.5µL`
`20µL` reaction: `7µL`
3. Incubate at `65°C` for `0h 10m 0s`
Program in PCR Machine: RT-DI1
4. Prepare the mix

5. Add mix to sample
`10µL` reaction: `3.5µL`
`20µL` reaction: `7µL`
6. Incubate at `50°C` for 60 minutes then `85°C` for `0h 5m 0s` then `4°C` forever
Program in PCR machine: RORT2SPE
7. Keep in `-20°C` for at least `0h 20m 0s` before moving on to the PCR
PCR
Same kit and steps for both cbVG and Genome
3/3) PCR
Procedure
1. Prepare master mix (can be done on bench) * Volumes below are for when `1µL` of cDNA is used. Adjust dH<sub>2</sub>O volume accordingly if more cDNA is added. Up to `4µL` cDNA can be added but increases background. Total reaction volume should equal `25µL`.

2. Add `24µL` of master mix to PCR tubes (can be done on bench) 3. Thaw cDNA samples and spin down in mini microcentrifuge 4. Add `1µL` of cDNA sample to master mix (add in PCR hood! cbVG are very stable!) 5. Run PCR
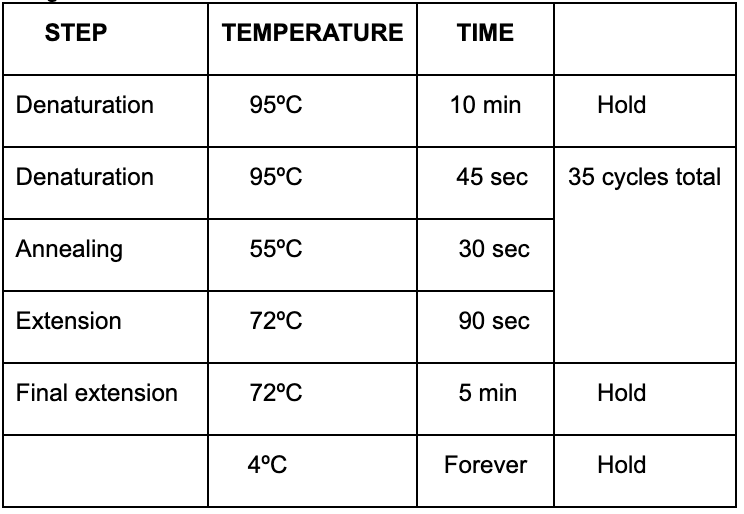
Program in PCR Machine: DIPCR
Gel Electrophoresis
1. Prepare 1% agarose gel
1 g pure agarose in `100mL` of 1X TAE buffer
Microwave until agarose solution dissolves completely (\~3 min)
Let agarose solution cool before adding Ethidium bromide (if you can keep your fingers on the
flask without burning, then it is at an appropriate temperature)
Add `1-5µL` of Ethidium bromide and mix by swirling flask
2. Pour agarose solution into gel cast (remember to put in the well comb)
3. Let the agarose solidify (wait at least `0h 30m 0s`)
4. Place gel in an electrophoresis chamber containing 1X TAE buffer (make sure buffer covers gel)
5. Load ladder and samples to wells
`6µL` of Ladder
Ladder stock recipe:
`100µL` Gene Ruler 100 bp Plus DNA ladder from Thermo Scientific: SM0321
`100µL` DNA Gel Loading Dye (6X) from Thermo Scientific: R0611
`400µL` dH<sub>2</sub>O
`30µL` of Sample + Loading dye mix (dilutes from 6X to 1X)
`5µL` of DNA Gel Loading Dye (6X) from Thermo Scientific: R0611
`25µL` of sample
6. Run at 120 volts for 30-40 minutes
7. Analyze gel bands
Sendai Virus Genomic and cbVG PCR CONCEPT
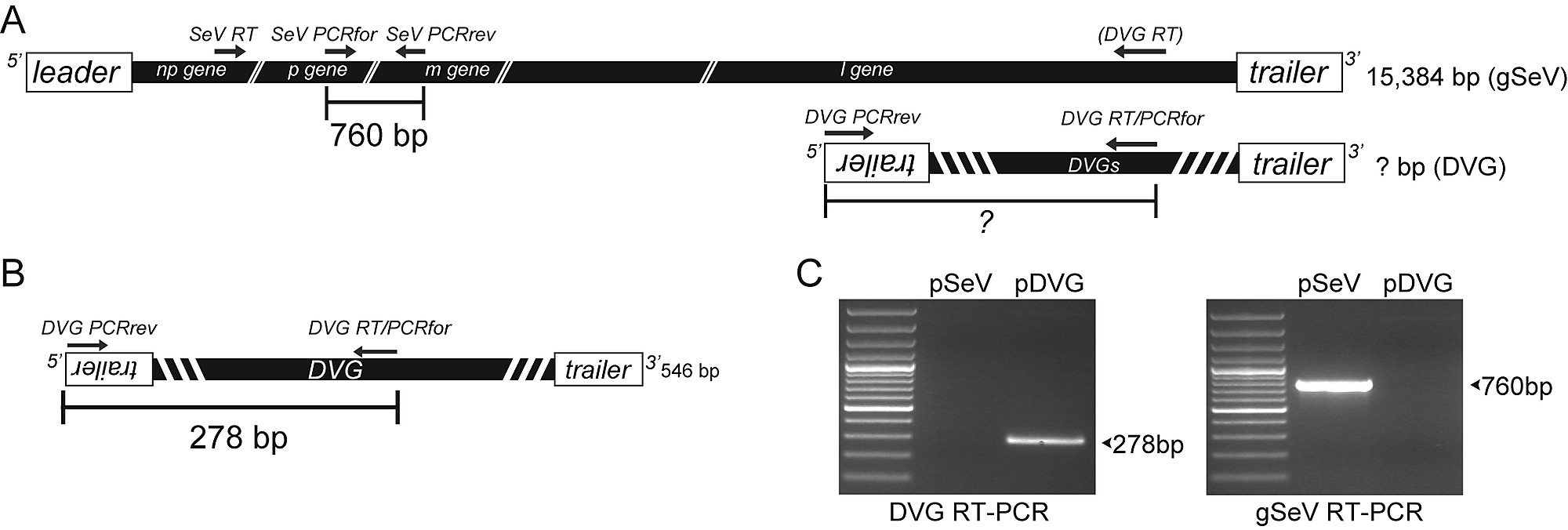
SeV copy back VG PCR strategy and validation. ( A ) Diagram of the genomic composition of the full-lengthSeV genome (gSeV) and of a representative copy-back VG of unknown length. Arrows indicate the location ofprimers used for RT and amplification (PCR). Full-length size of the genome is indicated. Expected amplicon size of 760 nt of the gSeV to be detected through our PCR assay is indicated. This strategy allows detection of most copy-back VGs replicating in an infected cell. ( B ) Schematics of SeV strain Cantell's predominant 546 nt long cbVG. Expected amplicon size of 278 nt of this particular cbVG to be detected through our PCR assay isindicated. ( C ) Validation of the cbVG PCR assay. cbVG and gSeV amplicons from plasmids encoding the full-length SeV strain Cantell genome (lane 1) or the SeV strain Cantell dominant cbVG (lane 2) after amplificationusing the primers depicted in (A). https://doi.org/10.1371/journal.ppat.1003703. https://doi.org/10.1371/journal.ppat.1003703.

