Salmonella detection with spin column extraction and 7500-FAST enrichment PCR
Laura Goodman, Rebecca Franklin-Guild, Renee Anerson
Disclaimer
Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
Abstract
This procedure is used to test enrichment broth from environmental or animal specimens using Qiagen spin columns and the MicroSEQ Salmonella spp. Detection Kit.
The RVS enrichment broth should be prepared in advance to allow for quality control. The rest of the method is performed over two days:
Day 1 - sample setup and enrichment
Day 2 - DNA purification and real-time PCR
Before start
Steps
Preparation of RVS broth and Quality Check
This broth is used for the enrichment of specimens for Salmonella spp. isolation. The Rappaport Vassiliadis (RVS) media has an expiration date of 6 months from the preparation date .
Determine batch size:
| A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|
| RO water | 0.5 L | 1 L | 2 L | 3 L | 4 L | 5 L |
| RVS broth powder | 13.55 g | 27.11 g | 54.22 g | 81.33 g | 108.44 g | 135.55 g |
RVS powder (Rappaport Vassalidius) can be obtained from HIMEDIA (#M1491)
Add half the amount of RO water needed for the batch and a sterile stir bar to a sterile
flask of appropriate size
Place the flask on a stir plate and start rotating the stir bar to “stir” the water
Add the entire amount of RVS powder to the flask while stirring
Add the remaining amount of water to the flask
Heat the flask until the powder is completely dissolved
Dispense 10 ml of the broth from the flask into sterile 20 x 125 mm screw-capped
tubes using sterile tubing and the DoseIt. Loosely cap each tube.
Autoclave the tubes of broth for 15 minutes on liquid cycle @ 121oC
Allow to cool to RT and then tighten caps.
Label racks of tubes as “RVS Broth”, date made, initials,& expiration date (months)
Perform QC using the fields below according to the manufacturer’s directions
Store RVS Broth @ 2-8oC protected from light
Quality Control Procedure:RVS Broth
RVS Broth/BP inoculated:Initials:Date:
BP examined:Initials:Date:
BP :□ growth □ no growth
BG/XLD inoculated:Initials:Date:
BG/XLD Subculture plates examined: Initials:Date:
Positive Control plates:
BG: □ typical colonies □ atypical colonies □ no growth □ mixed growth
XLD:□ typical colonies □ atypical colonies □ no growth □ mixed growth
Final Results(check one) Final Results (check one) Pass □ Fail/Batch Discarded □
Date:
Initials:
Sample processing and enrichment
Thoroughly disinfect a biosafety cabinet (BSC) using a disinfectant spray. Place a piece of absorbent bench paper in the BSC after cleaning. Allow the BSC to run for several minutes before placing items in it.
Obtain the following and place into the running BSC:
- Prepared aliquots of RVS (9ml per sample) to accommodate the total number of samples being tested plus one negative enrichment setup control. Positive controls are optional for this exercise. Retrieve additional aliquots of RVS if you will be adding positive controls.
- Racks to hold 15 ml tubes
- Lab towels soaked with 10% bleach contained within a plastic holder.
- Small biohazard bag (to collect waste)
- Labeled samples (organized in appropriate sized rack)
- Optional: one positive control culture each for Salmonella Typhimurium and Salmonella Dublin grown on blood agar
- Sterile 10 µL loop (optional if using an in-house positive culture control)
For the Vet-LIRN Salmonella Interlaboratory Comparison Exercise, samples to be tested will be liquid suspensions of bovine intestinal scrapings in PBS. The test sample tubes contain 1ml of sample in a 15 ml tube.
Place the first sample in a tube rack and loosen the cap. Clean your entire gloved hand with the 10% bleach towel.
Chose one 9ml RVS aliquot and pour the entire volume into the first sample tube. Use care to avoid back splashing. Recap the tube. Discard the empty aliquot tube in the biohazard waste.
Clean your entire gloved hand with the 10% bleach towel. Place the inoculated tube, which now has both sample and RVS broth, back into the original rack. Clean your gloves again.
Repeat steps 16-19 with the remaining samples.
Reserve one aliquot of RVS broth and label as negative setup enrichment control: this tube will remain un-inoculated. Note! After over-night incubation, the negative setup enrichment control will be extracted and tested by PCR along with the samples.
Optional: Inoculate one positive control using a sterile loop.
Move the rack(s) to a 40-44oC incubator, with shaking preferred. Incubate overnight, at least 18-24 hours.
DNA Extraction
Add 200 μL ethanol (200 proof) to each tube and mix thoroughly by vortexing followed by a brief pulse spin.
Transfer 1 mL of culture to a labelled microcentrifuge tube and centrifuge at 5,000 g for 10 minutes and remove supernatant.
Add 180 μL Buffer ATL to the tube and resuspend the pellet. Add 20 μL Proteinase K to the tube, then vortex.
If the ATL buffer solution is cloudy and has precipitate formation, be sure to heat the solution at 50 - 55°C until solution is clear before adding to culture.
Incubate at 56°C for a minimum of 1 hour or up to overnight, vortexing periodically.
Optional: Add 4 μL of RNase A to each tube, vortex and then incubate at RT for 3-5 minutes.
Add 200 μL Buffer AL to the tube.
Pipet the mixture from step 14 into a labelled DNeasy Mini spin column. Centrifuge at 6,000xg for 1 minute. Discard flow-through and the collection tube.
Add 500 μL Buffer AW1, and centrifuge for 1 minute at 6,000xg (8,000rpm). Discard the flow-through and the collection tube.
Transfer the spin column to a new collection tube.
Centrifuge for 1 minute at 18,000-20,000 x g (14,000 rpm) to dry.
Add 500 μL Buffer AW1, and centrifuge for 1 minute at 6,000xg (8,000rpm). Discard the flow-through and the collection tube.
Add 500 μL Buffer AW2, and centrifuge for 1 minute at 6,000xg (8,000rpm). Discard the flow-through and the collection tube.
Transfer the spin column to a new collection tube.
Centrifuge for 1 minute at 18,000-20,000 x g (14,000 rpm) to dry.
Place the spin column in a clean, labelled 1.5 mL microcentrifuge tube. Pipet 100 μL Buffer EB directly onto the DNeasy membrane.
Incubate at room temperature for 1 minute and then centrifuge for 1 minute at 18,000-20,000 x g (14,000 rpm) to elute.
Discard spin column and store DNA sample at 4°C for up to 48 hours. For long term storage (>48 hours), store DNA sample at -20°C.
PCR Setup
Program the ABI 7500-FAST machine with the following settings (use fast mode):
| A | B | C | D |
|---|---|---|---|
| c | Reps | Temperature | Duration (sec) |
| Stage 1 | 1 | 95 ͦ C | 20 |
| Stage 2 | 40 | 95 ͦ C | 03 |
| 60 ͦ C | 30 |
Set up reactions at ROOM TEMPERATURE (do not use cold blocks or ice). Chilled nucleic acid and lyophilized components will make resuspension of the reaction mixture difficult.
Slowly remove the rounded caps on the tube strips, one row at a time, and discard them.
Add 30 µl of sample nucleic acid elution to the PCR tube containing the lyophilized master mix. Wait 3-5 seconds and then resuspend the reagent pellet by pipetting up and down 2-3 times.
Important ! It is critical to wait 3-5 seconds to avoid aspirating part of the still lyophilized PCR master mix into the pipette tip.
Add 30 µl of the negative amplification control (supplied with the kit) to a PCR tube containing the lyophilized master mix. Wait for 3-5 seconds and then resuspend the reagent pellet by pipetting up and down 2-3 times.
Add 30 µl of the positive amplification control to a PCR tube containing the lyophilized master mix. Wait for 3-5 seconds and then resuspend the reagent pellet by pipetting up and down 2-3 times.
Cap the tubes with the flat caps provided with the kit. The cap strip must be seated level as shown in the picture.
Place a small mark at one end of each of the strip caps in order to keep proper orientation and order of the tubes. Do not mark the sides of the tubes or place any marks over the main surface of the optical cap.

Pulse spin the strip tubes in order to collect liquid in the bottom of the tubes and displace any air bubbles.
Place Samples in ABI-7500 FAST thermocycler using the tube strip adapter. Balance the tubes on each side according to the manufacturer’s instructions (blank tubes may be used for this purpose if necessary). If you do not have this adapter or are using a different machine, you can transfer the reactions into a plate that is compatible with your setup if needed.

Start the run.
Following run, analyze the results using the automatic analysis setting. All samples should have a VIC (internal control) signal. If the VIC is >35 or undetermined, repeat the PCR using a 1:5 dilution of the sample in nuclease-free water and interpret based on the result that gives the lowest Ct for the VIC.
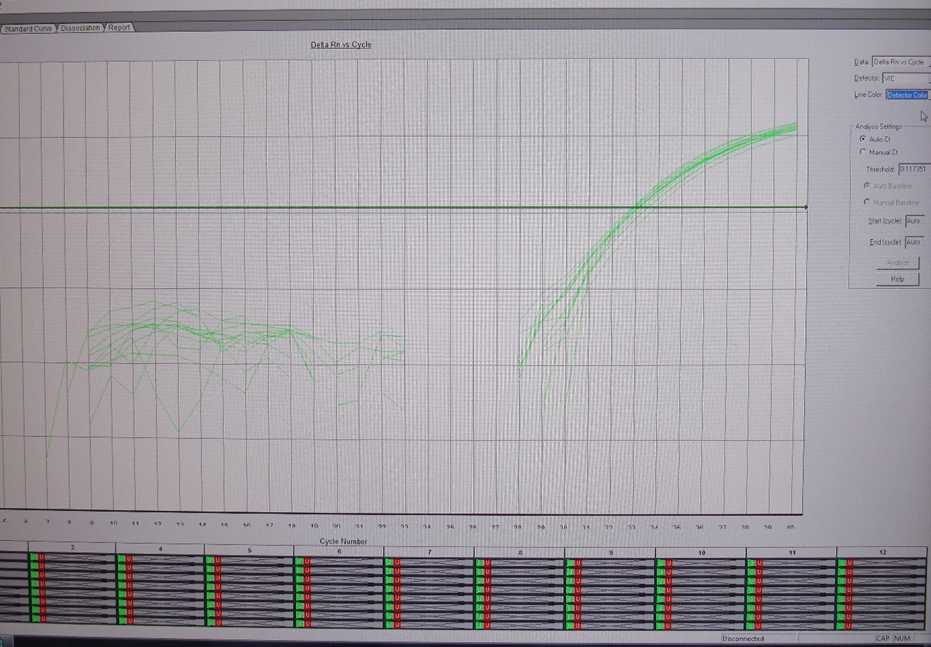
Save a copy of the original run file and export the data into an Excel file. Interpret the results according to the following guidelines:
| A | B | C | D |
|---|---|---|---|
| FAMa | VICb | Interpretation | Action |
| undetermined | detected | Not Detected | None – this result is final |
| < 35 | disregard | Positive | Culture confirmation is optional |
| ≥ 35 | detected | Suspect | Culture confirmation is optional |
| undetermined | undetermined | Inconclusive | Culture confirmation is optional |
aFAM (6-carboxyfluorescein) is the most commonly used fluorescent dye attachment for oligonucleotides and is calibrated as part of the standard set of dyes for the ABI 7500-FAST machine. Its max excitation wavelength is 495 nm and emission wavelength is 520 nmbVIC is an ABI-proprietary dye that with excitation wavelength 488 nm/Emission Wavelength 552 nm that is also calibrated as part of the standard set of dyes for the ABI 7500-FAST machine.
Copy the results into the provided SecureSheet file.
Optional: proceed with culture confirmation for samples with a Salmonella (FAM) Ct detected

