SARS-CoV-2 NCBI consensus submission protocol: GenBank
Ruth Timme, Emma Griffiths, Lee Katz, Duncan MacCannell, Michael Weigand
Disclaimer
Please note that this protocol is public domain, which supersedes the CC-BY license default used by protocols.io.
Abstract
This is a SARS-CoV-2 specific protocol that covers the steps needed to submit SARS-CoV-2 consensus sequence to GenBank.
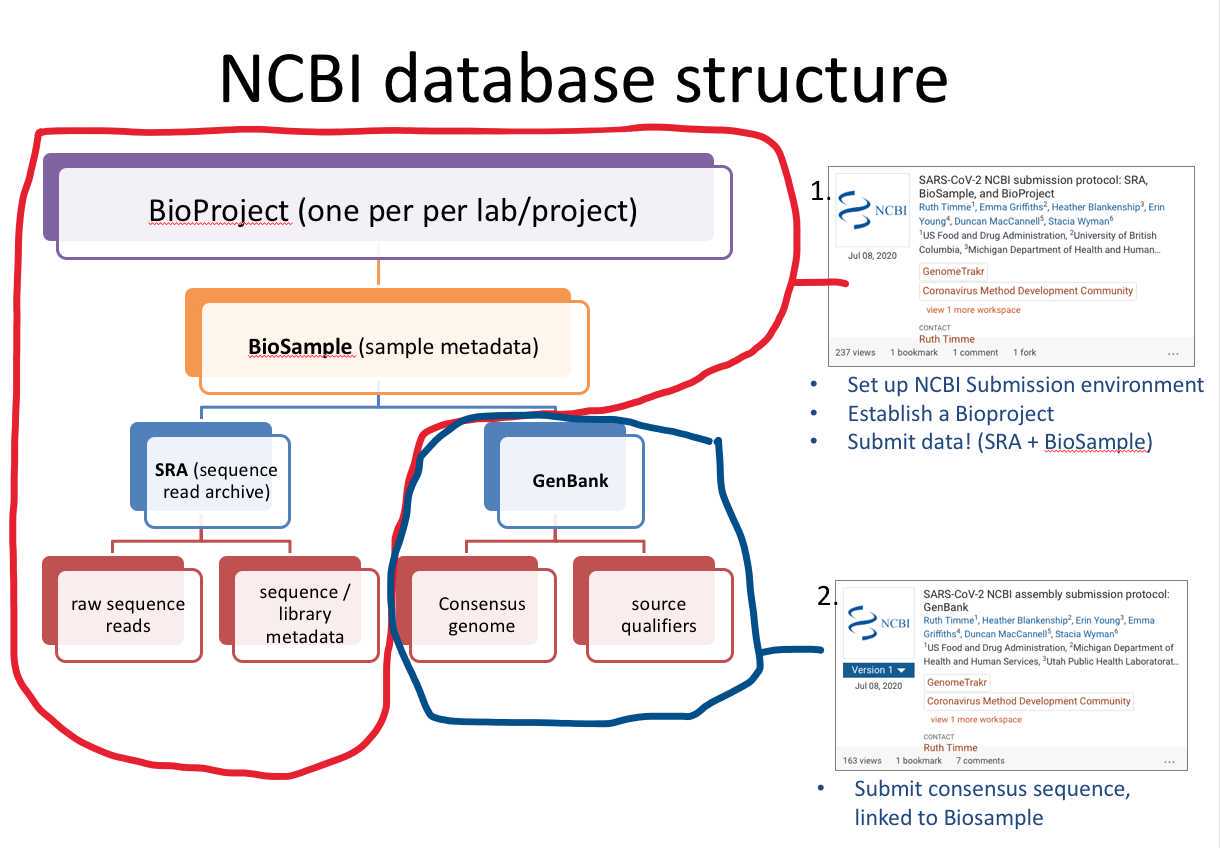
If you need a pipeline for frequent or large volume submissions, follow Step 1 in the SARS-CoV-2 NCBI submission protocol: SRA, BioSample, and BioProject SARS-CoV-2 NCBI submission protocol: SRA, BioSample, and BioProject to get your NCBI submission environment established, then contact gb-admin@ncbi.nlm.nih.gov to set up an account for submitting through the API.
This protocol assumes (and requires) that the user has a BioProject and BioSamples(s) already registered.
Complete in order::
1. Populate your templates first. Populate your templates first.
2. SARS-CoV-2 NCBI submission protocol: SRA, BioSample, and BioProject SARS-CoV-2 NCBI submission protocol: SRA, BioSample, and BioProject
- Step-by-step instructions for establishing a new NCBI laboratory submission account and for creating and linking a new BioProject to an existing umbrella effort.
- SARS-CoV-2 raw data submission to SRA (Sequence Read Archive) and metadata to BioSample. Users can modify this protocol to just create a BioSample with no linked raw data.
3. SARS-CoV-2 NCBI consensus submission protocol: GenBank (included protocol) SARS-CoV-2 NCBI consensus submission protocol: GenBank (included protocol)
_Required_ : established BioProject and BioSamples
- Submit SARS-CoV-2 assemblies to NCBI GenBank, linking to existing BioProject, BioSamples, and raw data.
Version history:
V3: Direct links provided to download metadata templates (instead of hosting duplicate files). minor edits throughout the protocol.
Before start
This protocol has two sections:
Section 1 : ensuring your NCBI submission environment is established
Section 2 : SARS-CoV-2 submission of assemblies or consensus sequences to GenBank.
Associated protocols:
- SOP for populating the NCBI submission templates (e.g. source modifiers for GenBank)
- NCBI submission to BioProject, SRA, and BioSample. Also includes NCBI account set-up for new users (Step 1)
- NCBI Data Curation protocol for making updates, corrections, or retractions to your data.
Link to PHA4GE contextual data specification PHA4GE contextual data specification
Steps
"Ingredients" to have in place before starting your submissions
1.1: Ensure you have a working NCBI user account
1.2 : Identify your NCBI submission user group or establish a new one if necessary.
1.3: Bookmark the link to your submission portal
1.4. BioSample + BioProject accessions in-hand
After these steps are complete you can proceed with GenBank data submission in Step 2.
Sign in to you r NCBI user account : https://www.ncbi.nlm.nih.gov/account/

Ensure you have an NCBI user group established and correct permissions are assigned for you to submit.
List of submission groups: https://submit.ncbi.nlm.nih.gov/groups/
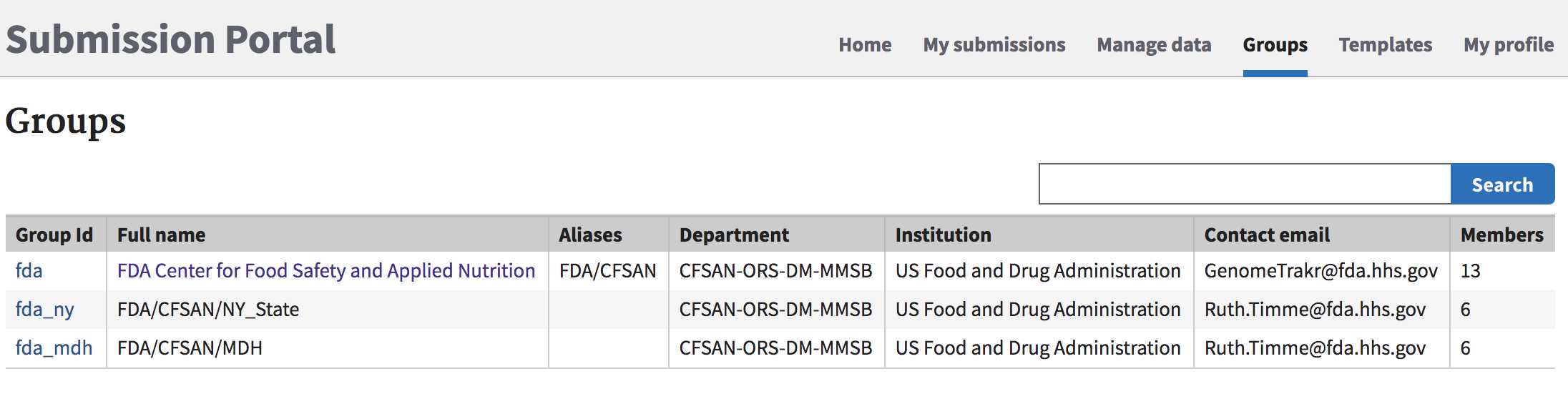
If you don't have a submission group established, please follow this protocol to create one for your laboratory group:
https://www.protocols.io/edit/sars-cov-2-ncbi-submission-protocol-sra-biosample-bf7bjrin
Bookmark “my submissions” at NCBI: https://submit.ncbi.nlm.nih.gov/subs/. This is your homepage for tracking your NCBI submissions.
If you see a blank page with a yellow box in the upper right corner saying “please login”, click this link and login using the credentials created in Step 1.1 .
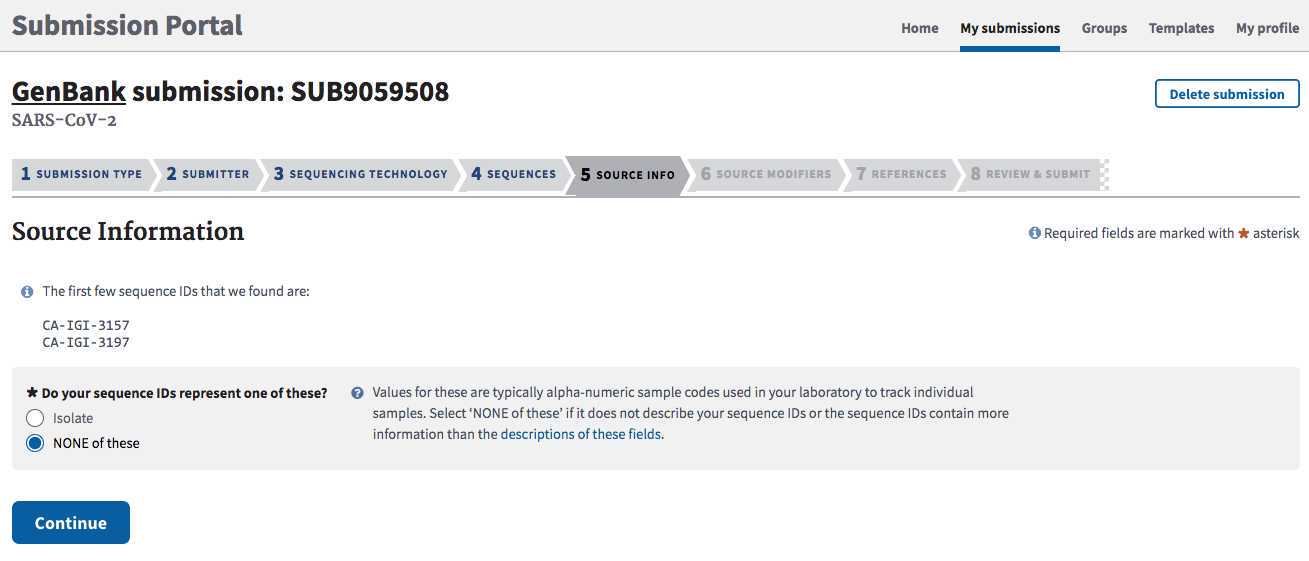
Identify your lab's BioProject accession. Does your laboratory have an established BioProject for this effort?
If not please follow instructions in Step 3 of https://www.protocols.io/edit/sars-cov-2-ncbi-submission-protocol-sra-biosample-br8ym9xw for creating a new one.
Data submission (assemblies to GenBank)
GenBank consensus (or assembly) submission of SARS-CoV-2:
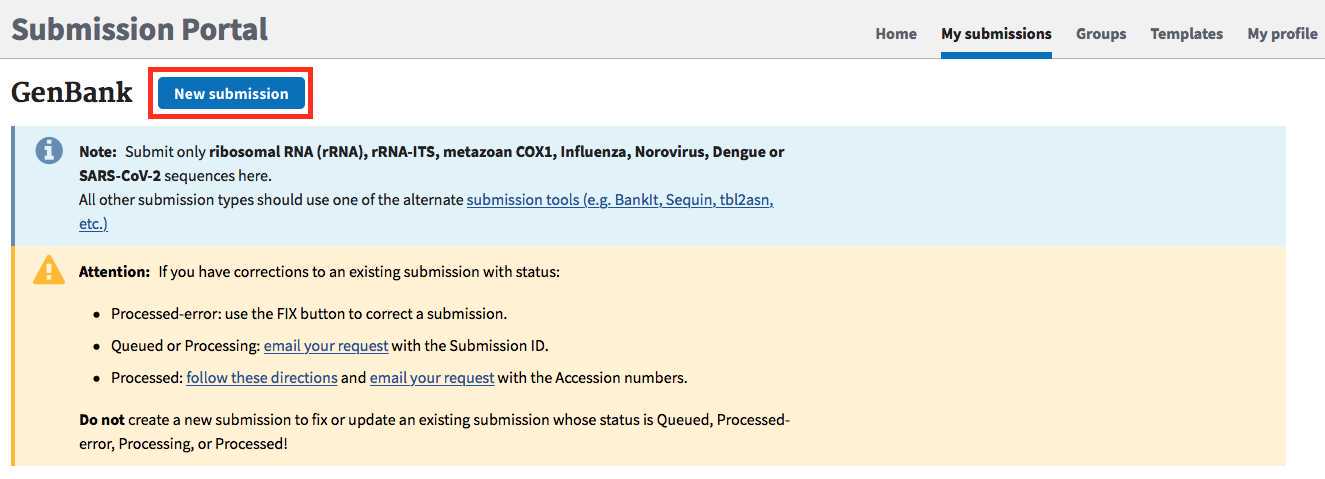
SARS-CoV-2 landing page: https://submit.ncbi.nlm.nih.gov/sarscov2/
Click "submit" under GenBank.
Populate the two GenBank templates.
Download the current templates in Step 4 Overview of NCBI's SARS-CoV-2 submission process and the metadata required.
- GenBank structured comment info (mapping/assembly methods)
- GenBank Source Qualifiers
Populate the metadata spreadsheets for each isolate you intend to submit (you can submit metadata for a single isolate, entire MiSeq run, or for a large collection of isolates you intend to submit in batch).
Save the source qualifier excel spreadsheet as a tab-delimited text file (.tsv) and ensure that the date field is formatted correctly (e.g. 2020-04-20) in the text file.
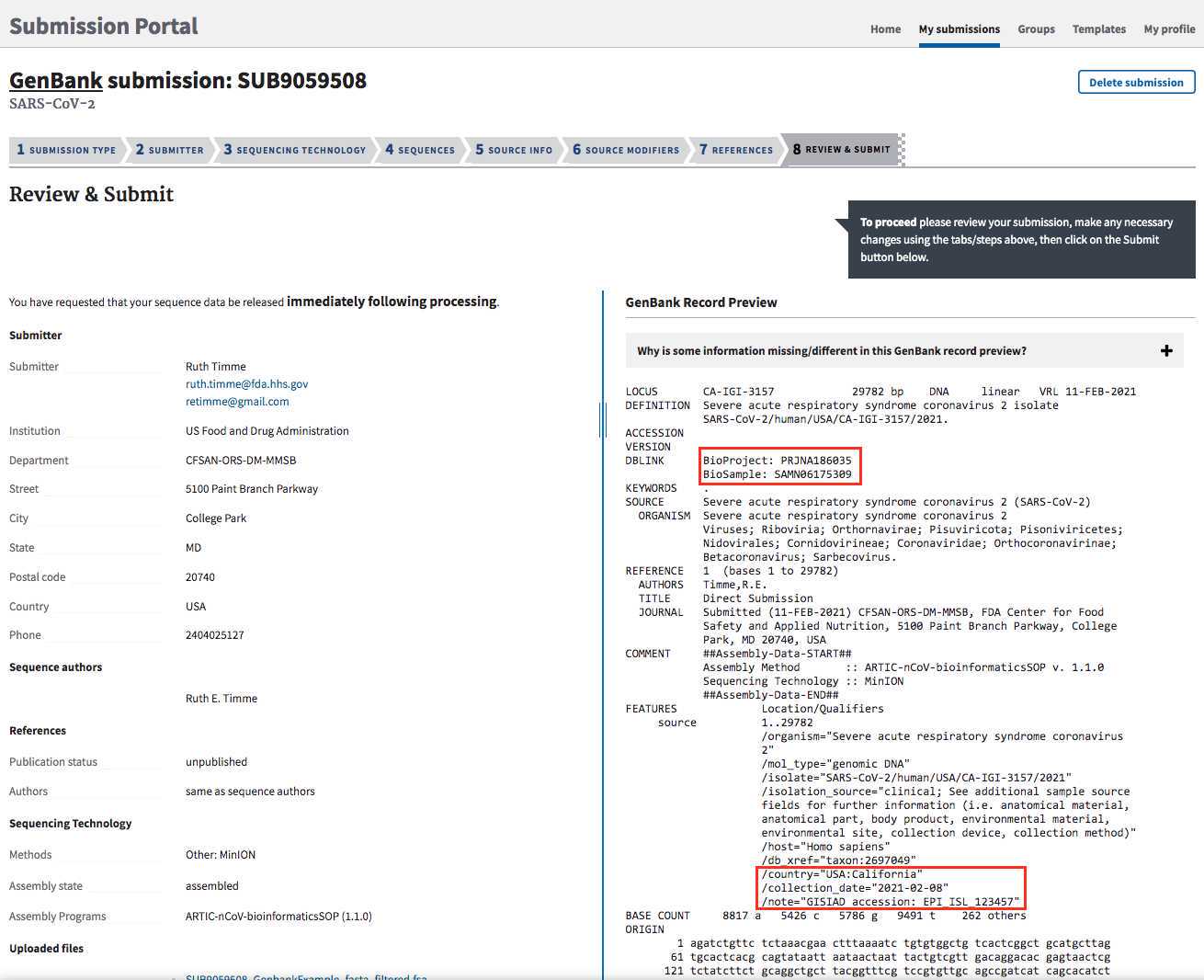
GenBank accessions:
The status of your submission will be updated once it is processed and can be tracked within the "My Submissions" tab: https://submit.ncbi.nlm.nih.gov/subs/.
GenBank accessions will be listed here, under "GenBank: Processed" and available for download in the AccessionReport.tsv file.
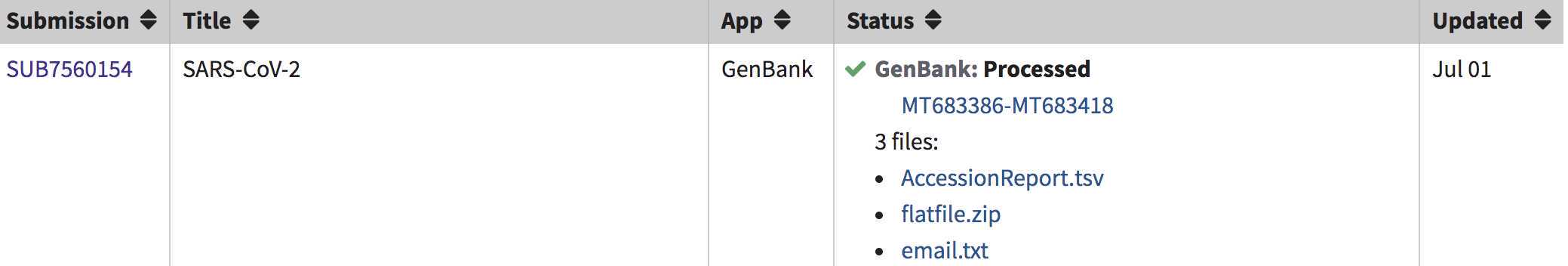
Sequences with no annotation issues will be listed as Processed. Submissions with annotation discrepancies will be marked as Error and a Fix button will appear. A report is emailed to you and listed on the submissions page with the detailed issues. If the data is incorrect, click the Fix button and you will return to the sequences page of your submission to upload a corrected file.
If you have evidence that the discrepancy is due to a naturally occurring mutation, send an email to gb-admin@ncbi.nlm.nih.gov with the SUB number and evidence.
Important data stewardship and curation notes:
- Develop an internal method for storing and tracking your GenBank accessions! They are required for making future updates to your records.
Prepare your sequence files.
Concatenate all SARS-CoV-2 consensus sequences into a single fasta file, where the fasta headers contain the "sample_name" submitted to the BioSample.
Example FASTA file for two sequence submissions:
CA-IGI-0042
ATCGATCGGTACCTAAGGATCGATCGGTACCTAAGGATCGATCGGTACCTAAGG....
CA-IGI-0031
ATCGATCGGTACCTAAGGATCGATCGGTACCTAAGGATCGATCGGTACCTAAGG....
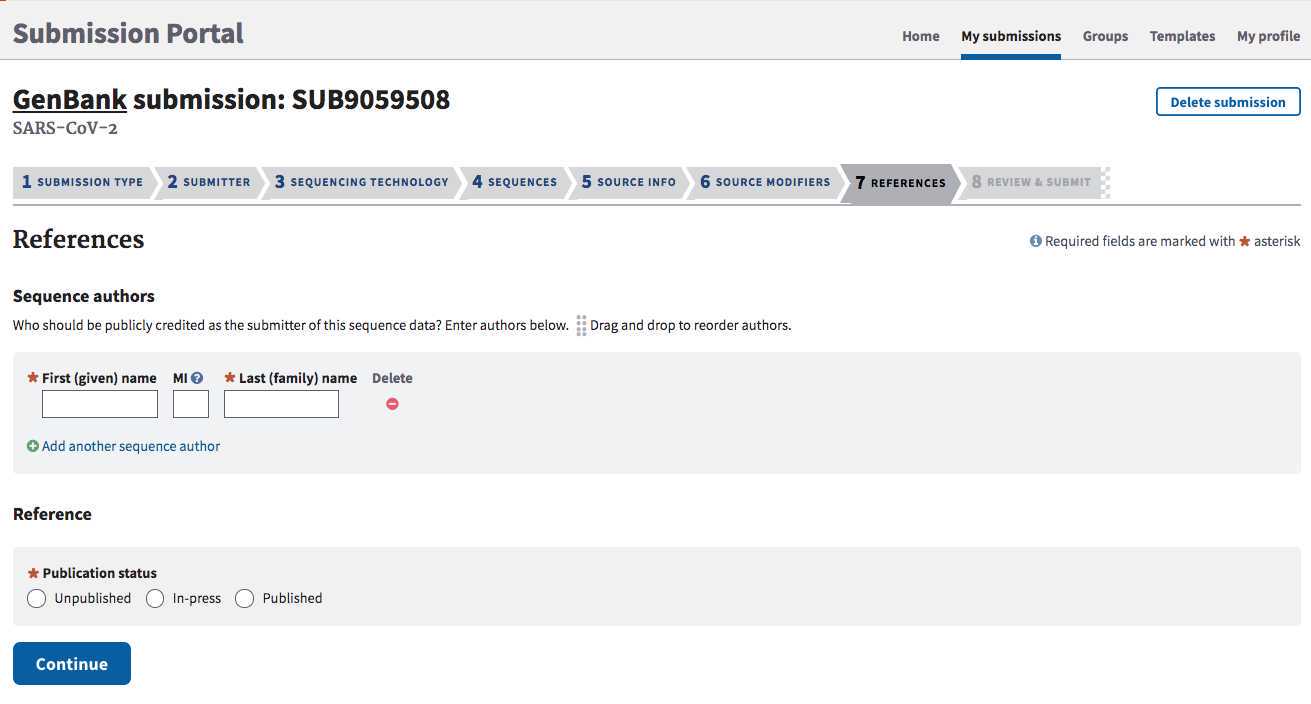
SUBMITTER tab:
Populate with submitter info. The “submitter” is the name of the person, or user group, who is physically doing the submissions, not a supervisor or PI.
**Must be the same person or group that submitted the associated BioSamples and BioProject.
Select the appropriate submission group name for your laboratory and check the contact information below.
**If you do not have a submission group available to click, see Steps 1.2-1.3 in the SRA submission protocol to establish a new one for your laboratory, or to add your name to a group already established for your lab.
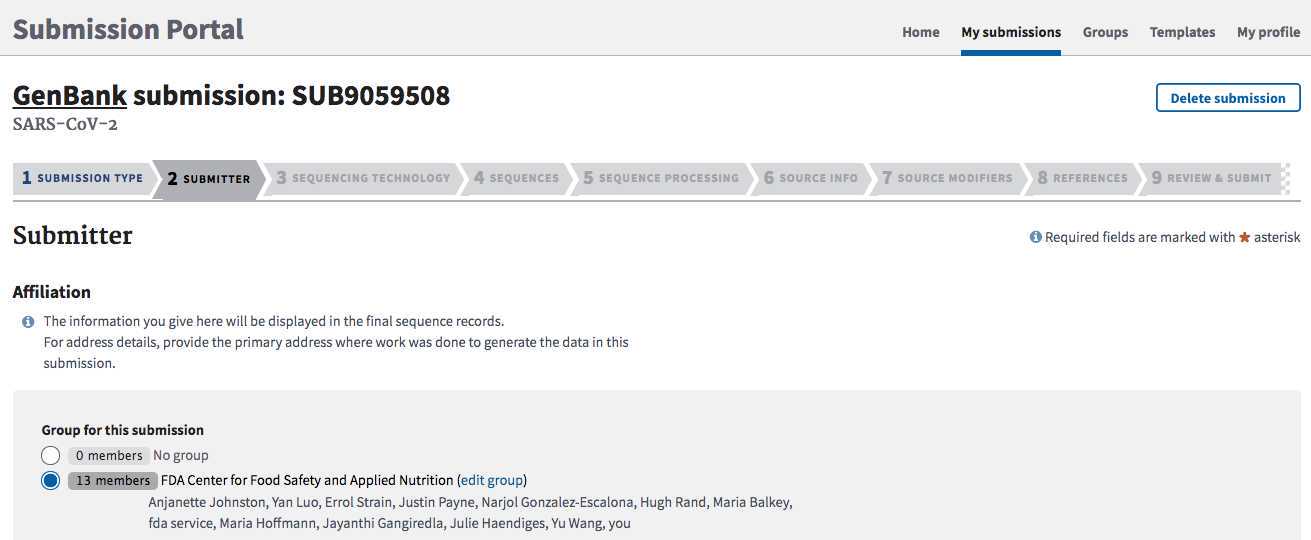
Click "Continue" to proceed.
SEQUENCING TECHNOLOGY tab:
This information will get populated as a structured comment on the GenBank record.
Pull the sequencing method and assembly information gathered in Step 2.1.
Method : sequencing technology or platform.
Assembly State: Click "Assembled sequences".
Assembly information : Specify program/pipeline AND version.
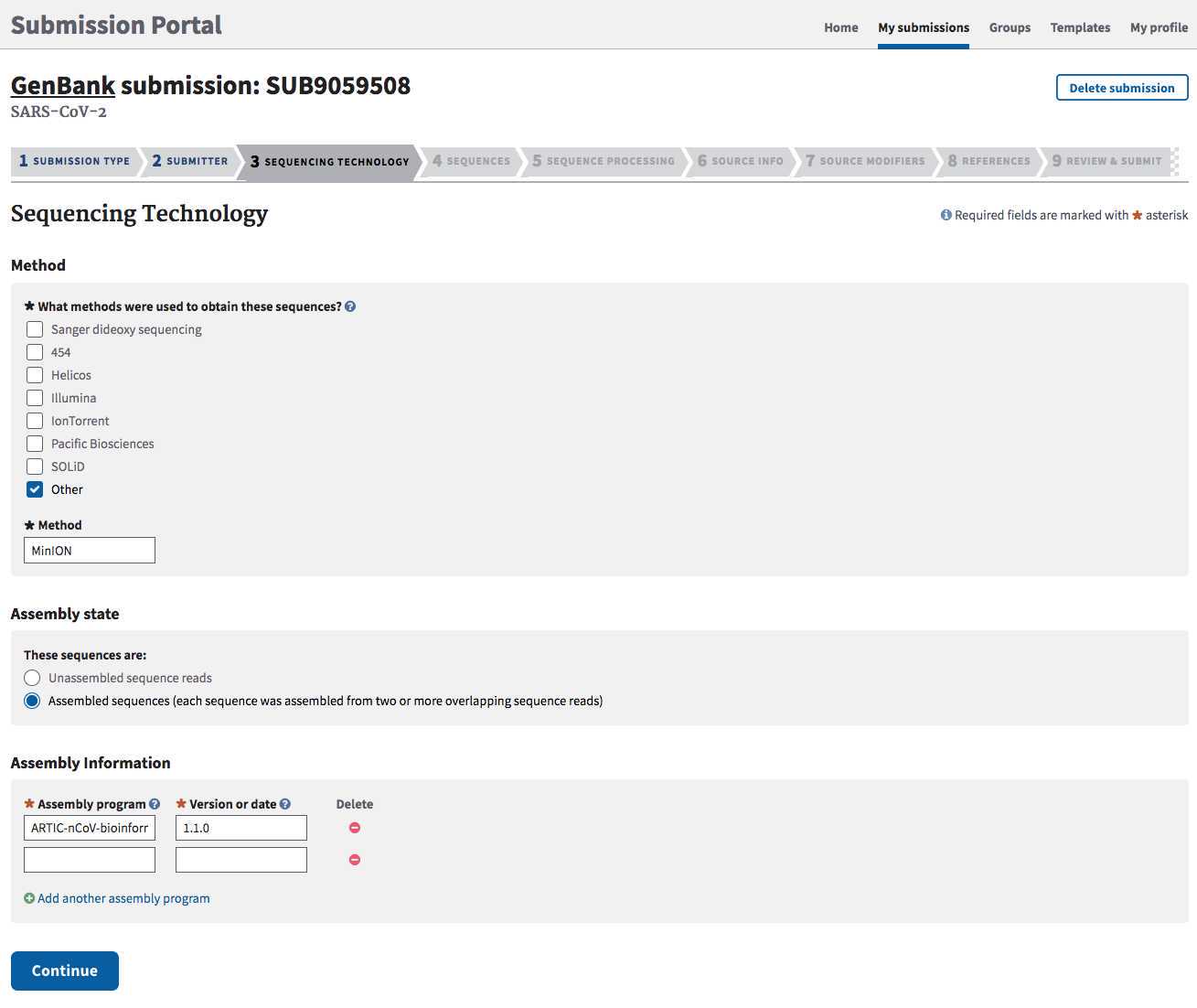
Click Continue to proceed.
SEQUENCES tab:
Release date: Click "Release immediately following processing" for all routine surveillance isolates.
Sequences:
Sequences can be uploaded one at a time (one per submission), or as a batch upload in a single concatenated FASTA file (https://submit.ncbi.nlm.nih.gov/genbank/help/#fasta) when you are submitting multiple isolates in one submission. See Step 2.1 for guidance on formating your FASTA file.
Click "Choose File" to browse and upload your .fasta file:
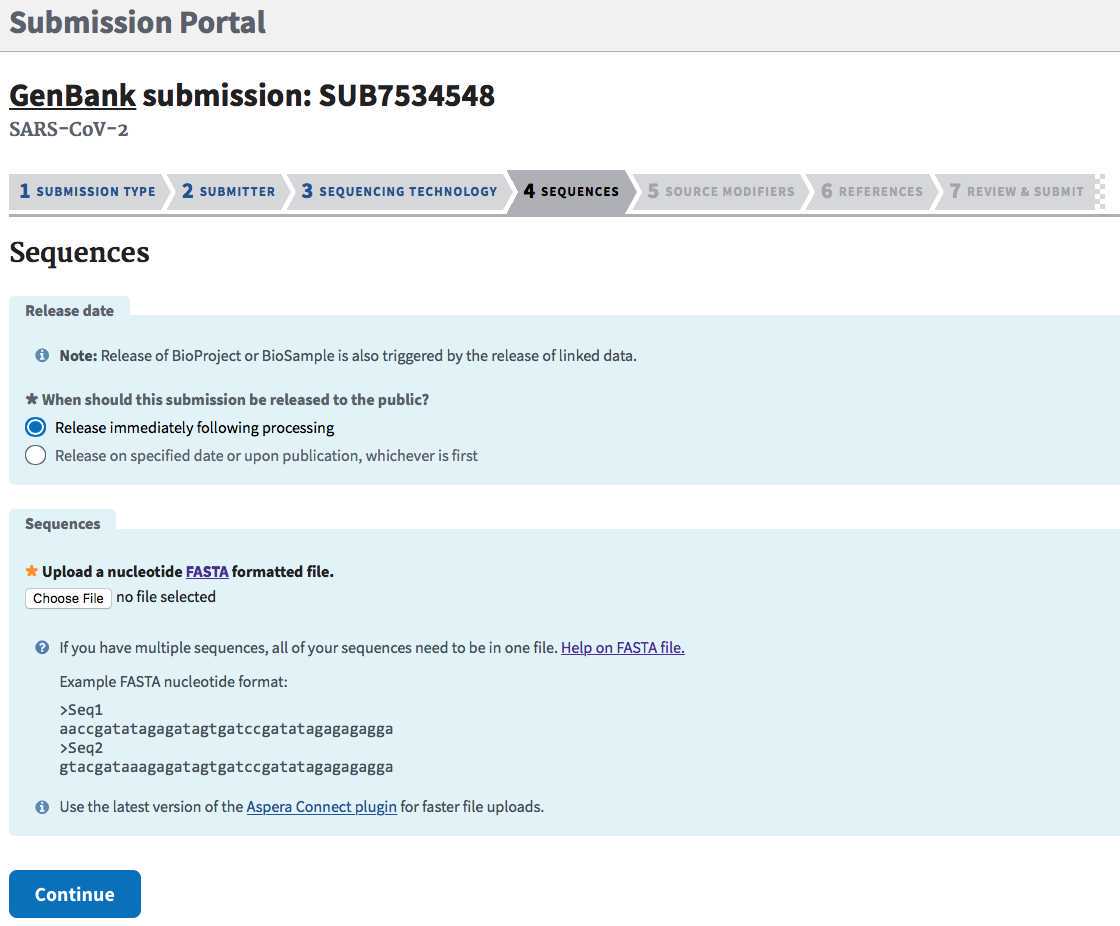
Click "Continue" and respond to any validation issues.
Common validation issues:
Ambiguous bases were trimmed warning. Ambiguous bases are non- A, T, G, or Cs. NCBI trims terminal Ns first at the 5' end, then looks to see if 50% of the first 10 bases are ambiguous and trim to last ambiguous. If more than 30% of the first 50 are ambiguous, we trim to the last ambiguous and then recheck the 5' end. If that is fine, we follow the same steps on the 3' end. This procedure is run again if we trimmed vector from an end. NCBI removes sequences that are greater than 50% ambiguous after the trimming. They also remove sequences with internal vector.
String of NNNs : If your assembly contains strings of internal NNNs (from mapping to a reference genome), you will get a warning asking for you for more information:
-
Click "A region of estimated length between the sequenced regions based on an alignment to similar sequences or genome" if the NNNs were caused by the reference-based assembly.
-
Click "Release immediately following processing".
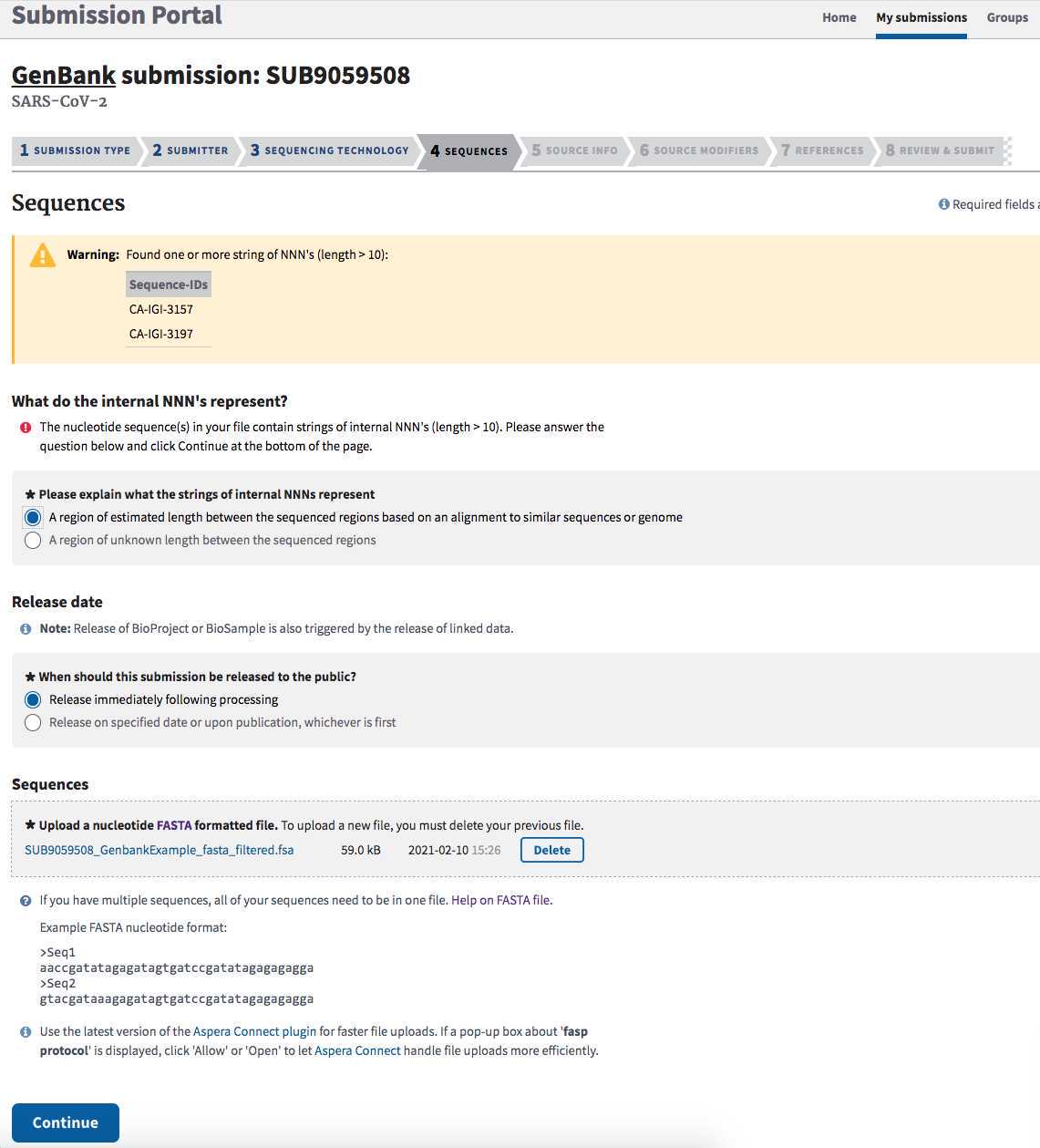
Click "Continue" again.
SOURCE MODIFIERS tab:
Guidance for populating this metadata outlined in Step 2.2.
- Click "Upload a tab-delimited table"
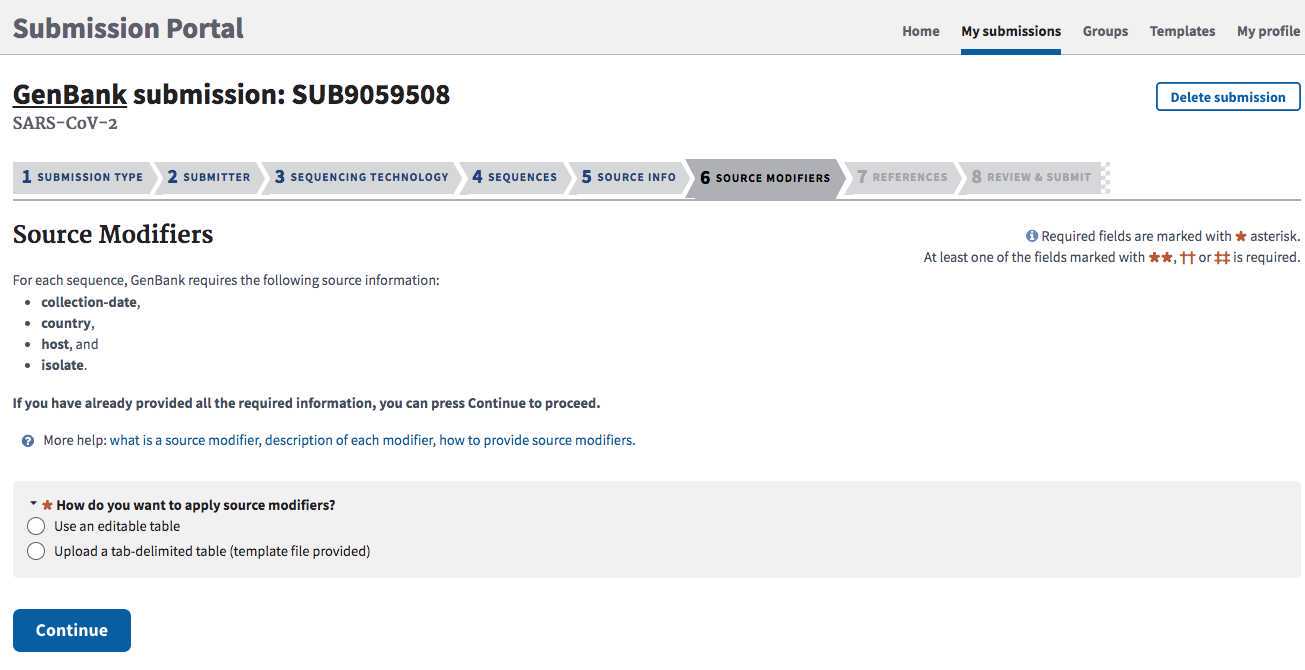
Upload the csv file created from populating the PHA4GE GenBank source modifiers template in Step 2.2. Upload this file by clicking on the "upload a tab-delimited text file" link. Ensure that the first column in this spreadsheet, "Sequence_ID" contains an ID that matches exactly the ID used in your FASTA file headers.
Click Continue .