SARS-CoV-2 Infection and Viral Replication of Human Lung Organoids
Morris Baumgardt, Stefan Hippenstiel, Andreas C. Hocke, Katja Hönzke, Maren Hülsemann, Karen Hoffmann, Mirjana Kessler
Disclaimer
Informed written consent was obtained from all volunteers and the study was approved by the Charité Ethics Committee (project 451, EA2/079/13).
Abstract
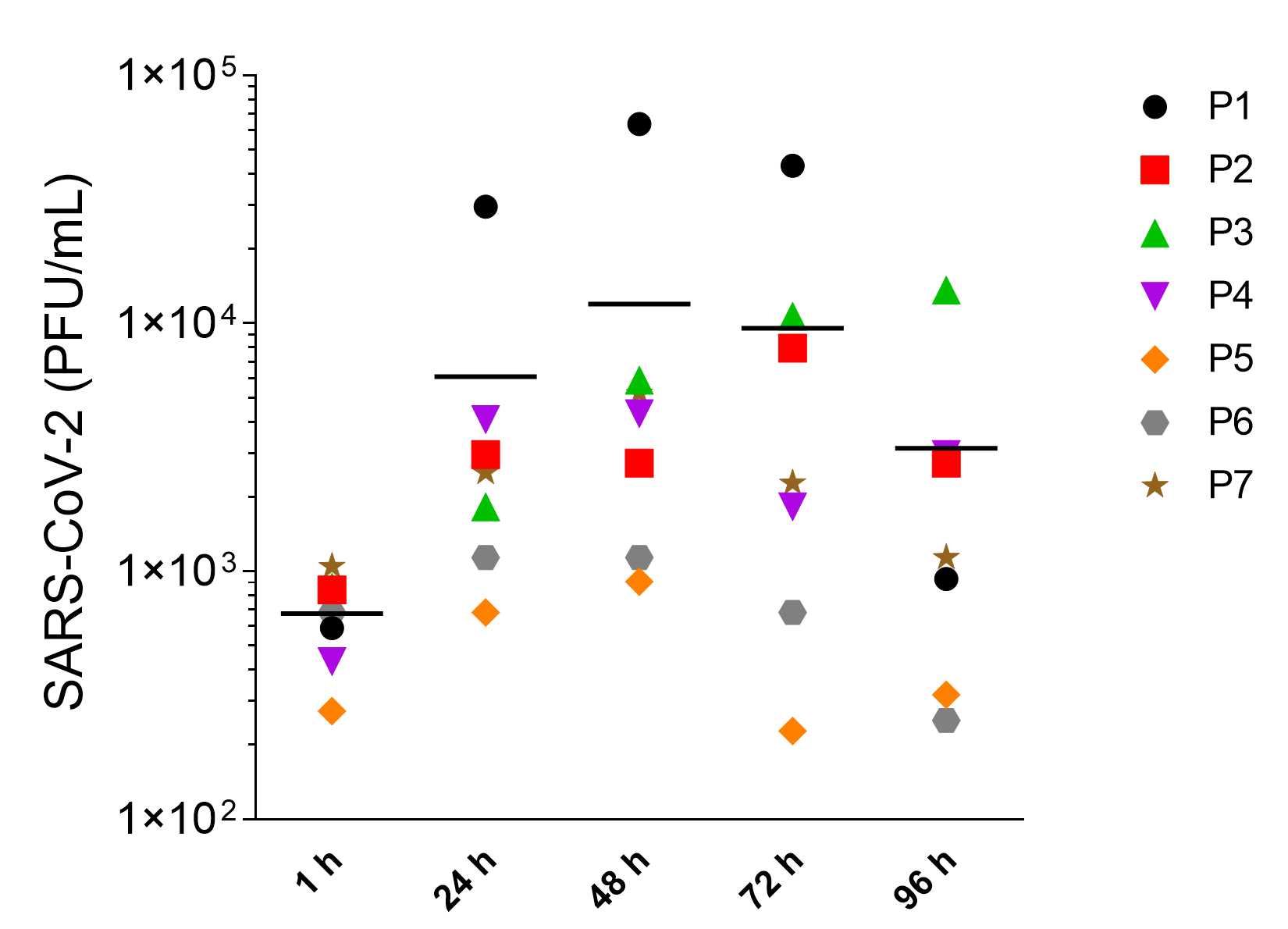
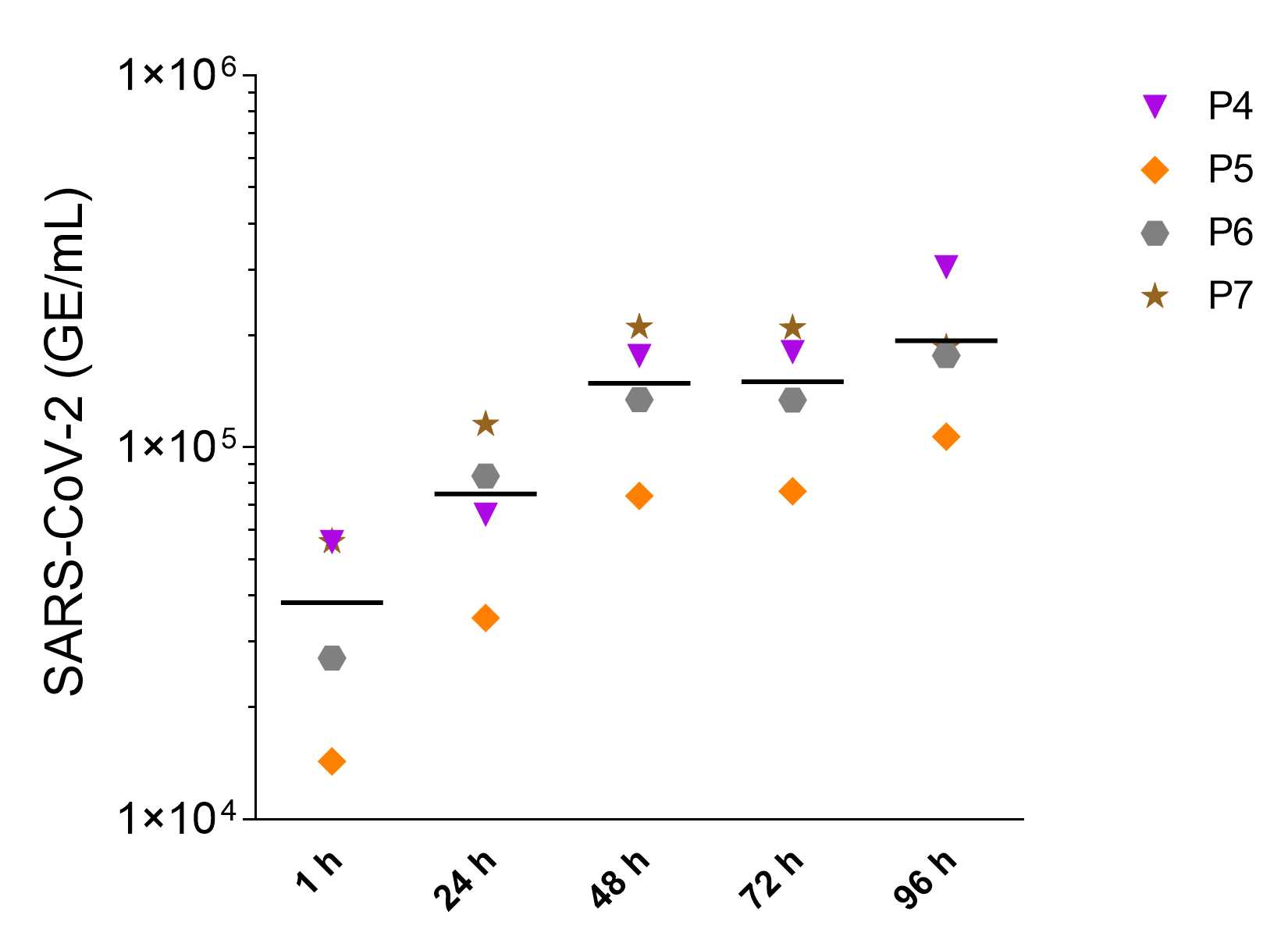
This protocol describes the working steps to infect human alveolar-like organoids with SARS-CoV-2 and quantify the viral replication at defined time points post infection via plaque assay and viral E gene quantitative reverse transcription PCR (RT-qPCR).
Before start
Grow the virus stock (SARS-CoV-2 B.1) on Vero E6 cells (RRID:CVCL_0574), please work with maximum passage 3 and sequence the virus stock initially.
One day before the plaque assay will be performed, Vero E6 cells need to be seeded in order to ensure confluency and readiness for the plaque assay the next day. Therefore, 175,000 cells per 24-well are seeded in 500 µL Vero E6 medium. The next day, verification is made to ensure confluency of the cells.
Steps
Cultivation of human alveolar-like organoids
Grow your 3D model as described
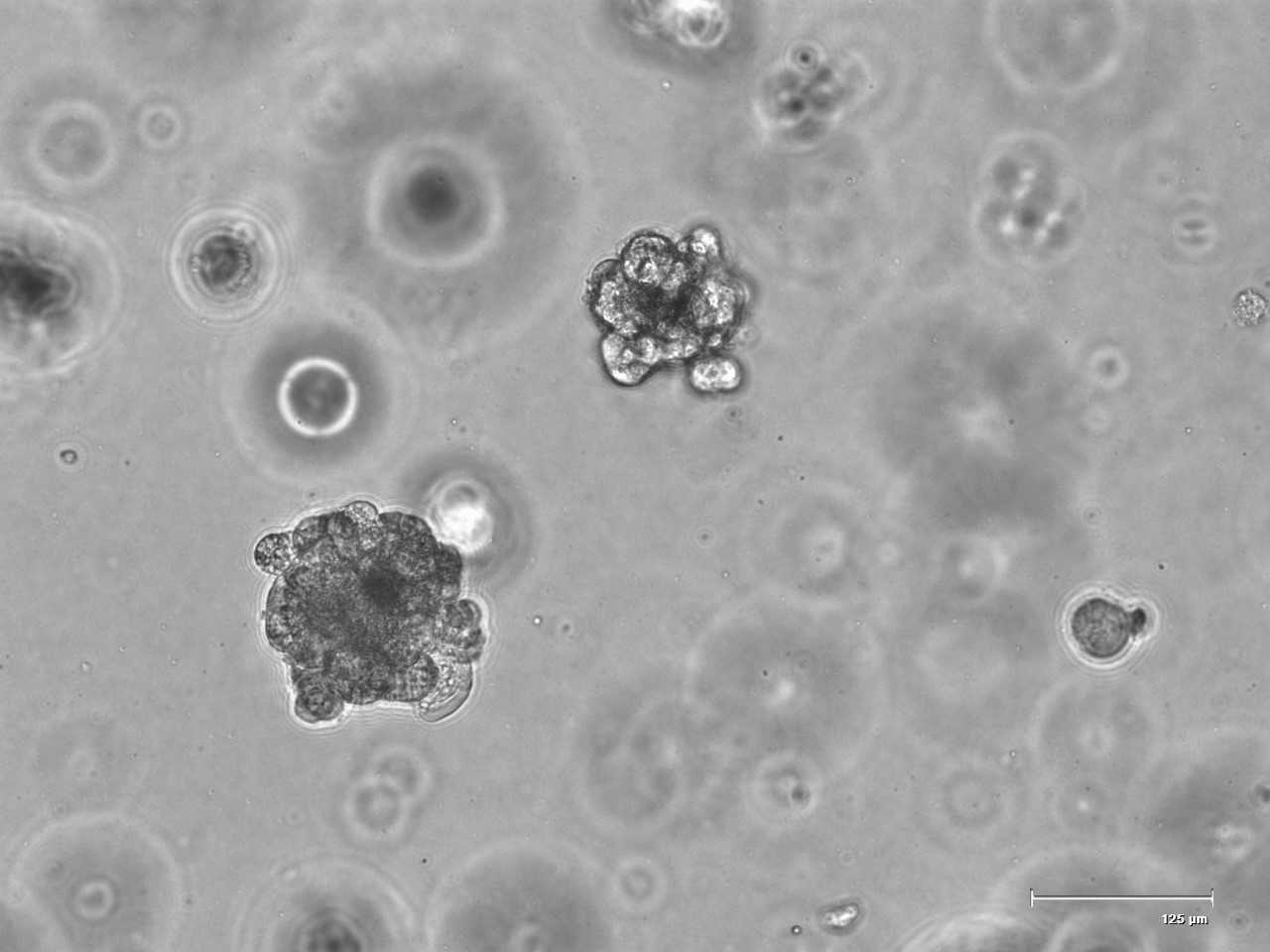
Cell counting of human alveolar-like organoids
A specific cell number is needed to calculate the virus MOI (multiplicity of infection).
Carefully remove organoid medium of one well.
Add 1mL (4°C) cold base medium and collect Cultrex with organoids in a 15mL tube, flush well with additional 1mL base medium.
Centrifuge at 300x g,4°C and carefully remove supernatant.
Resuspend pellet in 1mL TrypLE Express for 0h 15m 0s in a shaking wather bath (37°C ) and add 1mL base medium afterwards.
Produce a single cell solution by resuspending three times with a disposable syringe with needle (27G).
Count single cells using a disposable Counting Chamber (Neubauer improved).
Infection of human alveolar-like organoids
Carefully remove organoid medium.
Add 1mL (4°C) cold base medium and collect the suspension containing Cultrex and organoids in a 15mL tube, flush well with additional 1mL cold base medium.
Centrifuge at 300x g,4°C.
Carefully remove supernatant.
Resuspend pellet in 1mL cold base medium.
Pipette virus solution (stock) to the organoids (MOI = 1), mix by pipetting (1mL total volume per 15 mL tube) and incubate 1h 0m 0s at 37°C. Shake the tube gently every 15 mins.
MOI calculation:
(Cell number per well x MOI)/virus stock titer = µL virus solution to be added per well.
Centrifuge at 300x g,4°C.
Transfer 50µL from the supernatant for input titer control (for viral qPCR later) in a new 1.5mL tube and freeze at -80°C at BSL3 (biosafety level 3) until further processing.
Take 100µL from the supernatant for input titer control (for Plaque Assay later) and mix 100µL supernatant with 100µL gelatin medium in a new 1.5mL tube and freeze at -80°C at BSL3 (biosafety level 3) until further processing in the plaque assay.
Carefully remove and discard as much remaining supernatant as possible from the original tube so that only oganoids remain.
Wash organoids with 500µL PBS (1x).
Centrifuge at 300x g,4°C.
Incubate 0h 30m 0s at 37°C.
Plaque Assay
Samples for plaque titration are taken from the organoid culture supernatant at different time points/hours post infection (hpi) as described here:
Pipette Cultrex and medium vigorously up and down (using 1000 µL pipette) in the well until the Cultrex is homogeneously dispersed in the medium and transfer Cultrex, organoids and virus containing medium into a 1.5mL tube.
For the complete dissolution of the Cultrex and thereby remove all organoids and viral particles from the matrix, cool the tube with organoid mixture for 0h 10m 0s at 4°C using a pre-cooled cooling rack.
After 0h 10m 0s at 4°C centrifuge at 300x g,4°C.
Take 100µL from the supernatant at the desired time points (e.g. input (0 h), 1 h, 24 h, 48 h, 72 h, 96 h) and mix 100µL supernatant with 100µL gelatin medium in a new 1.5mL tube and freeze at -80°C at BSL3 (biosafety level 3) until further processing. At the same time, freeze 50µL of pure supernatant (without gelatin) in another 1.5mL tube, as these will be used later for viral RT-qPCR (-80°C).
Make sure, that Vero E6 cells are confluent before infection (see "Before start").
Thaw the obtained supernatants of infected organoids from step 28 and prepare the following dilution serie:
Prepare 10-fold serial dilutions:
Mix 50µL of thawed supernatant and 450µL OptiPro in a 1.5mL tube by vortexing (closed cap, moderate power, ~0h 0m 5s) to receive a dilution of 10-1. Mix again 50µL of the 10-1 dilution with 450µL OptiPro in a new tube to receive a 10-2 dilution and continue this procedure until the desired dilution is achieved. (In the here described condition for infection a supernatant dilution of not exceeding 10-5 is adequate for the detection of SARS-CoV-2 replication in alveolar-like organoids).
Infect the Vero E6 cells: (add 200µL/24-well in duplicates using the desired dilutions).
Exemplary dilutions which can be used:
Input (necessary dilution depends on the virus titer): 10-1, 10-2, 10-3
1 hpi: 10-1, 10-2, 10-3, 10-4
24 hpi: 10-2, 10-3, 10-4, 10-5
48 hpi: 10-2, 10-3, 10-4, 10-5
72 hpi: 10-1, 10-2, 10-3, 10-4
96 hpi: 10-1, 10-2, 10-3, 10-4
Incubate the plate at 37°C for 1h 0m 0s.
Remove supernatant and wash the wells once with PBS.
Mix 2.4% Avicel and 2xDMEM in a ratio of 1:1 and add 500µL Avicel overlay to each well.
Incubate at 37°C for 72h 0m 0s.
Remove Avicel overlay and wash wells once with PBS.
Inactivate the entire 24-well plate by adding 6% PFA for 0h 30m 0s in a biotainer and then export from the BSL3.
Remove plates from the biotainer and led them air dry.
Add crystal violet working solution and incubate for 0h 15m 0s.
Remove crystal violet working solution.
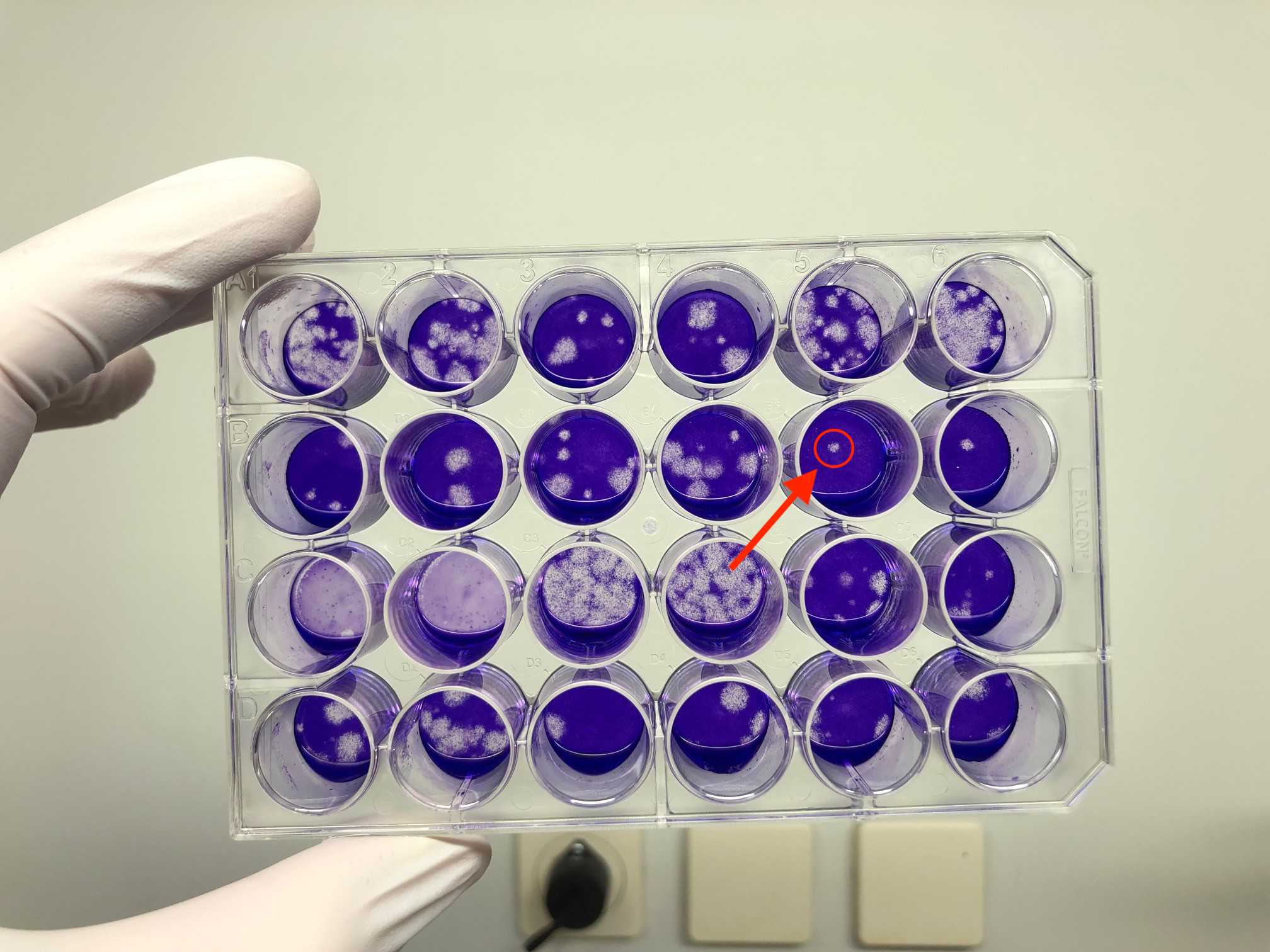
Document number of plaques in a table and take a picture of the stained plate.
Calculate PFU/mL (plaques from two dilution levels and two replicates are counted).
-
Sum of plaques given by the identified virus dilutions = Sum of plaques from 4 wells (e.g. 10-3= 2 wells, 10-4= 2 wells).
-
Titer of SARS‐CoV‐2 (in PFU/mL) = Number of plaques given by the identified virus dilutions ÷ (lowest dilution factor × 0.44 mL). Calculation is done with the to the lowest dilution referred inoculum volume of 0.44 mL * lowest dilution level.
For example :
10-3 dilution: 34 and 37 plaques/well = 71 plaques/two wells
10-4 dilution: 1 and 4 plaques/well = 5 plaques/two wells
Sum of all counted plaques / total volume of virus
--> 76 PFU/0.44 mL * 103 = 173103 = 1.73105 PFU/mL (the factor 103 corresponds to the lowest dilution level that was counted).
viral RT-qPCR
Samples for the quantification of SARS-CoV-2 RNA in supernatants of infected organoids were taken as described in step 28 . The samples were taken at different timepoints after infection and frozen.
Thaw the frozen 50µL supernatant samples at RT.
Pipet 50µL of virus containing supernatant into 300µL of RAV1 buffer with Carrier RNA (as instructed by manufacturer) and vortex.
Heat-inactivate the tubes at 70°C for 0h 10m 0s in a thermomixer (600rpm,0h 0m 0s) and export from BSL3.
Freeze the samples at -80°C or directly process them for qPCR by first isolating the viral RNA using Nucleospin RNA Virus Kit according to the manufacturer’s protocol.
Quantify the viral RNA copies from your samples ( viral qPCR Master Mix table: template RNA ) using Platinum SuperScript lll One-Step RT-PCR system with Platinum Taq DNA polymerase kit by preparing the master mix shown in the table below.
| A | B | C |
|---|---|---|
| Concentration | µL | |
| PCR-grade H2O | 1.8 | |
| 2X Reaction Mix | 6.25 | |
| MgSO4 | 50 mM | 0.2 |
| E_Sarbeco_F | 10 µM | 0.5 |
| E_Sarbeco_R | 10 µM | 0.5 |
| E_Sarbeco_Probe | 10 µM | 0.25 |
| SSIII/P.Taq enzyme Mix | 0.5 | |
| Total Volume Master Mix | 10 | |
| Template RNA | 2.5 | |
| Total | 12.5 |
viral qPCR Master Mix
Standards for E gene-based quantitative reverse transcription PCR: SARS-CoV-2 E gene (1x105 copies/μL) as used for clinical diagnostics. In vitro-transcribed control RNA for the E gene assay can be acquired through the European Virus Archive platform (www.european-virus-archive.com).
Prepare 104, 103 and 102 copies/μL from the standard stock using nuclease-free water and use four dilutions (105-102 copies/μL) for qPCR analysis. Caution! avoid contamination of test reaction with the highly concentrated standards.
Negative control: PCR-grade, nuclease-free H2O.
Evaluation is performed automatically using the Roche LightCycler Software.
| A | B | C | D |
|---|---|---|---|
| Stage | Temperature | Duration | Repetitions |
| 1 | 55°C | 10 min | 1 |
| 2 | 95°C | 3 min | 1 |
| 3 | 95°C | 15 s | 45 |
| 4 | 58°C | 30 s | 45 |
Thermal cycling qPCR program