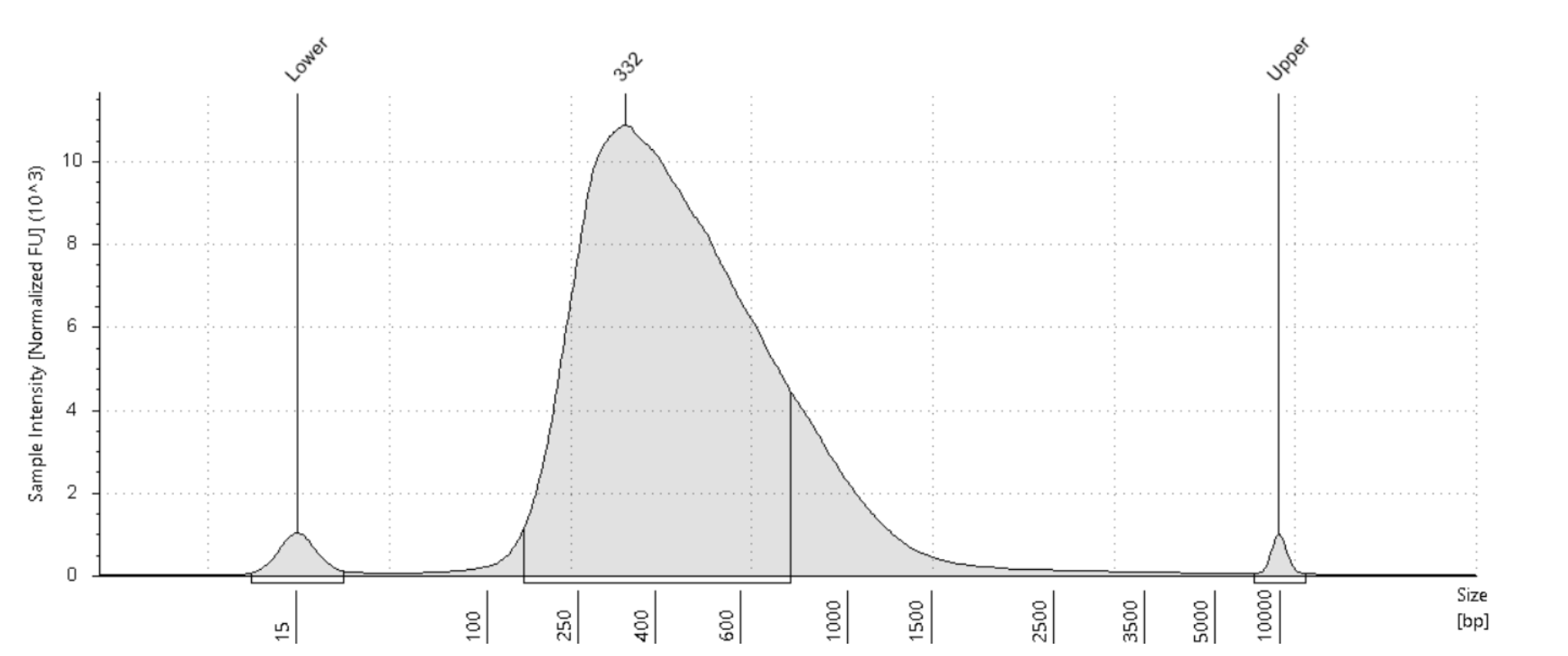
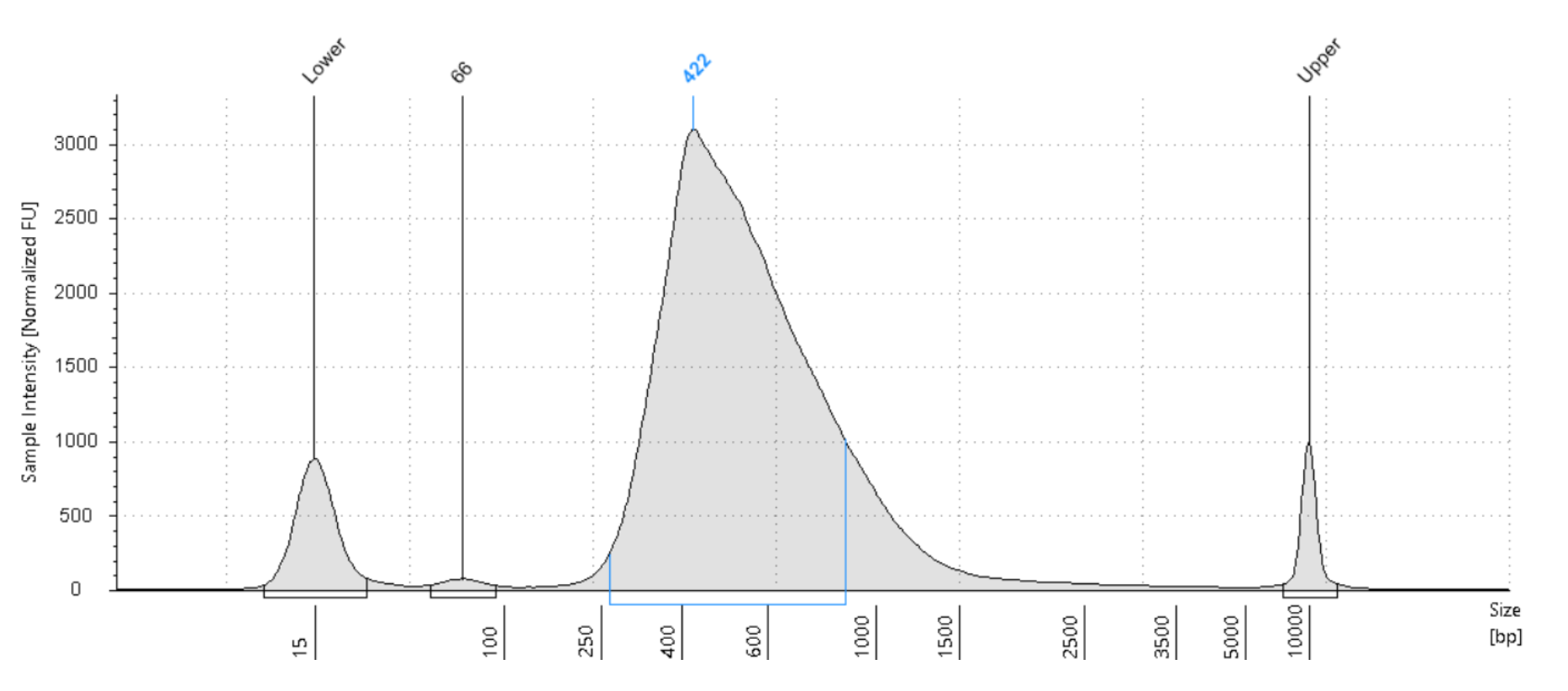
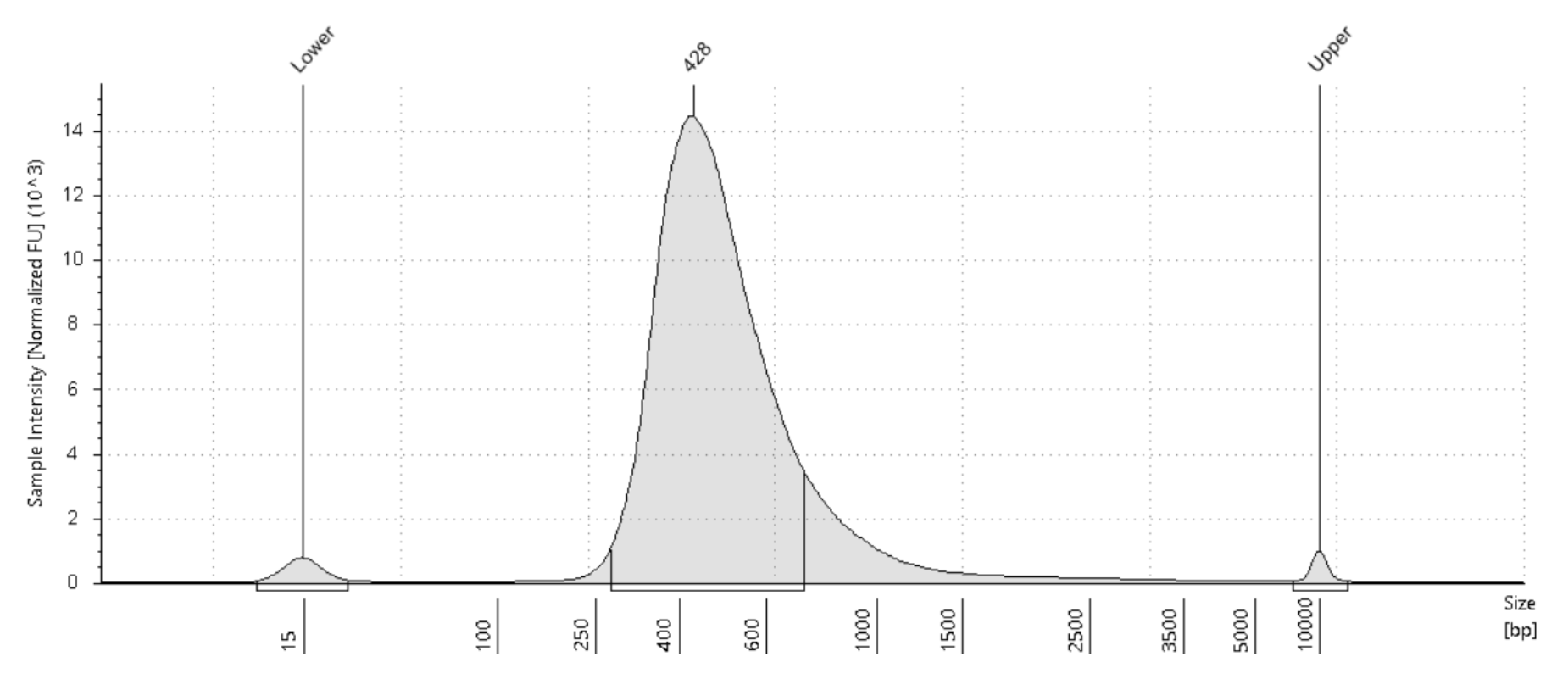
RoCK and ROI: bead modification, library generation and sequencing protocol
Giulia Moro, Konrad Basler, Erich Brunner
scRNAseq
targeted scRNAseq
capture
region of interest
whole transcriptome
BD Rhapsody
single cell sequencing
target
low expression
point mutations
targeted capture
splice junctions
Abstract
Various tools have been developed to reliably identify, trace and analyze single cells in complex tissues. In recent years, these technologies have been combined with transcriptomic profiling approaches to explore molecular mechanisms that drive development, health, and disease. A remaining challenge is that important information relevant for understanding the biology of cells or tissues, such as lowly expressed transcripts, sequence variations or exon junctions, remains undetected. We developed an scRNAseq workflow, RoCK and ROI (Robust Capture of Key transcripts and Region Of Interest), that tackles these limitations. RoCKseq uses targeted capture to enrich for key transcripts, thereby enhancing the detection, identification and tracking of cell types in scRNAseq experiments. ROIseq directs a subset of reads to a specific region of interest via selective priming. This allows specific sequence information to be retrieved for mRNAs of interest, enabling, for example, the inspection of sequence variations. Importantly, the targeted information obtained with RoCK and ROI is recorded together with standard transcriptome readouts. To analyze the multimodal information provided by RoCK and ROI, we developed a novel pipeline. The entire workflow increases the information obtained for lowly expressed genes and enables the detection of individual sequence variations and the exploration of the biological relevance and consequences of the respective variation for the cells expressing it.
This protocol covers the following steps:
-
Design of RoCKseq capture sequences and ROIseq primers
-
RoCKseq bead modification on BD Rhapsody beads
-
RoCK and ROI library generation
-
Sequencing of RoCK and ROI libraries
Before start
Important points to keep into consideration during RoCKseq bead modification
-
LoBind DNA tubes and pipette tips guarantee low bead loss during modification, which otherwise get stuck on walls of pipette tips and tube
-
Beads should be kept on ice whenever possible
-
Bead modification should be performed in a clean, RNAse-free hood
-
Enzymes should be kept at -20°C as long as possible and buffers and splints should be placed on ice after thawing
-
If multiple samples are processed in parallel, only wash up to four samples at a time to prevent incubation on the magnetic stand for too long
-
Try limiting (i.e. restrict to 1 minute) the time the beads are exposed to the magnetic stand
-
Avoid the drying out of the beads after washing
-
To minimise bead loss during modification: consistently use LoBind DNA Eppendorf tubes and LoBind pipette tips and wait for all the beads to be gathered at the magnet of the magnetic stand before exchanging buffers. During washes ensure that all liquid is expelled from the tip as to minimize bead loss
Important points to keep into consideration during the Fluorescent assay
-
After addition of the lysis buffer keep beads at room temperature. Do not place back on ice. This may lead to higher fluorescent background signal in the negative control
-
The fluorescent probes and the beads with the fluorescent probe should be kept in the dark whenever possible
Steps
Design of capture sequences
Before proceeding with the bead modification step, splints and fluorescent oligo need be designed and ordered
Points to keep into consideration when designing splints:
-
The GC content of splints should be in the range of 40-60%. Higher GC content may impair reverse transcription (i.e. first strand synthesis). Also consecutive GC stretches of more than 4 bases should be avoided. Similarly a low GC content and longer stretches of A should be avoided in order to prevent dT-based capture of the target transcript
-
GC content upstream of splint: if the GC content of the transcript of interest upstream of the splint is too high (more than 5 consecutive G or Cs), this may impair reverse transcription (i.e. first strand synthesis)
-
Length of the splint: 24 nucleotides
-
Place the capture whenever possible into the CDS of the transcript of interest: the 3’- and 5’ UTRs are less conserved and thus more prone to accumulate nucleotide polymorphisms that will hamper targeted capture. For long non-coding RNAs we suggest capturing the transcript in a conserved region whenever possible. Sequencing the locus in the strain used is recommended.
-
Vicinity to ROIseq primer: when performing RoCK and ROI, the splint should be chosen not more than 300 - 400 bp downstream of the ROIseq primer. This accounts for the sequence on the bead (primer, barcode, UMI, TSO). Please note adaptors for sequencing add to the final product size as well.
-
G or a C at the 5’ end of the splint (and thus 3’ end of the capture) favor reverse transcription.
-
The capture should not be overlapping with known splice junctions: this may be an issue if unknown splice variants are present (i.e. intron retention)
Splint sequences
IMPORTANT: : all splints are 5' phosphorylated
The sequence of the splint for the modification of TSO oligos on BD Rhapsody “Enhanced Cell Capture Beads V2” is as follows:
5’ -24 or 25 nt coding sequence followed by a constant sequence-3’:
5’-NNNNNNNNNNNNNNNNNNNNNNNNCATACCTACTACGCATA-3’
where the CATACCTACTACGCATA is the reverse complement of the TSO sequence on the beads.
The polyA protective oligo used on the barcoded beads is 18 nucleotides in length:
5’-AAAAAAAAAAAAAAAAAA-3’
The oligos should be ordered in 0.2 µmol scale, HPLC grade, with 5’ phosphorylation . Before use, resuspend the oligos in ddH2O to generate a 100 µM stock solution.
IMPORTANT:
-
To modify RoCKseq beads with multiple capture sequences, mix the splints in the desired ratio. For example, to modify RoCKseq beads with the same amount of three splints (33% each), pipette
5µLof each splint and mix with15µLof 100 µM polyA oligo -
The modification of RoCKseq beads can be titrated to achieve different amounts of modification on TSO oligos. The titration is achieved by mixing the splint(s) with the protective TSO oligo. This oligo is also 5' phosphorylated. For example, to achieve a 50% of RoCKseq modification, a mix of
7.5µLof splint(s) and7.5µLof protective TSO oligo is generated and mixed with15µLof 100 µM polyA oligo
Design of fluorescent oligos
To design the fluorescent oligos, take the first 20 nucleotides from the 5’ end of the splint .
The fluorescent oligos should be ordered in HPLC grade and in 0.2 µmol scale with a 5’ Atto647N modification and diluted in ddH2O to generate a 100 µM stock solution.
RoCKseq bead modification protocol for splint testing
IMPORTANT: the protocol described below refers to the modification of a full vial of BD Rhapsody barcoded beads. Alternatively, to test the efficacy of the bead modification with new capture sequences, the protocol can be adapted to modify a small aliquot of beads.
Instead of 2mLof BD Rhapsody barcoded beads per sample, 20µLof beads can be used. The same protocol can be used with the following changes:
Step 9: T4 polymerase mix: 40µL buffer, 20µL10 mM dNTPs, 136µL ddH2O
Step 28: 1µL of polyA – splint mix
Step 31: 1µLT4 polymerase enzyme
Step 34: lambda exonuclease mix: 15µL reaction buffer, 132µL ddH2O
Step 40: 3µL lambda exonuclease enzyme
The fluorescent assay protocol can be used as described below, with all 20µL of modified beads being used as input.
Step 1 modification of full vial of RoCKseq beads: preparation of reagents
Thaw lambda exonuclease buffer, T4 polymerase buffer, 100 µM splint(s), polyA oligo and 10 mM dNTPs at room temperature and place On ice
Preheat two thermomixers to 75°C and to37°C, respectively
Prepare TE/TW and Water buffers in 50 mL Falcons and place On ice
- TE/TW buffer:
500µLTris,100µLEDTA,10µLTween20, up to 50 mL with ddH2O - Water buffer:
10µLTween20, up to 50 mL with ddH2O
Preparation of T4 polymerase mix : Prepare four 1.5 mL DNA LoBind tubes. Pipette into each tube: 260µL T4 polymerase buffer, 130µL 10 mM dNTPs, 857µLddH2O and place On ice
Preparation of splint mix : Pipette 15µL of 100 µM polyA oligo and 15µL of 100 µM splint into new 1.5 mL DNA LoBind tube. If a mix of splints is used, pipette 15µL of 100 µM polyA oligo and 15µLof mix of splints (see Step 3)
Incubate splint mix in thermomixer at 75°C for 0h 5m 0s without shaking and place On ice
Preparation of beads : Resuspend the beads by gently pipetting up and down with a 1 mL pipette set to 500µL being careful not to lose any supernatant. Immediately transfer the2mL of barcoded beads provided by the manufacturer by pipetting 500µL of BD Rhapsody barcoded beads into four new 1.5 mL DNA LoBind tubes and place On ice. After the transfer to each tube resuspend the remaining beads by pipetting up and down to allow for a similar amount of beads being transferred per replicate
Proceed immediately to “Washing BD Rhapsody beads”
Step 2 modification of full vial of RoCKseq beads: washing BD Rhapsody beads
Place the four 1.5 mL DNA LoBind tubes containing the beads on a 1.5 mL magnetic stand
Wait until liquid in tubes is clear, takes about 0h 1m 0s to complete
Gently remove supernatant with 1 mL pipette without disturbing the beads - the LoBind tube remains on the magnetic stand
Remove first tube from magnetic stand and resuspend beads in at least600µL Water buffer, gently pipette up and down at least 5 times to resuspend beads and place the tube On ice
Repeat Step 17 with the other three tubes
Place the four 1.5 mL with washed BD Rhapsody beads on 1.5 mL magnetic stand
Repeat from Step 14 with TE/TW buffer processing one tube at the time as before
Resuspend the beads in at least600µLTE/TW buffer and place On ice
Step 3 modification of full vial of RoCKseq beads: T4 polymerase elongation
Place the four 1.5 mL tubes with washed BD Rhapsody beads on 1.5 mL magnetic stand and wait until liquid is clear, takes about 0h 1m 0s to complete
Remove supernatant from first tube - the tube remains on the magnetic stand
Resuspend beads from first tube with T4 polymerase mix (from Step 4) by gently pipetting up and down at least 5 times
Place the tube on a rack (non magnetic) at Room temperature
Repeat Steps 23-25 with the remaining three tubes
Mix splint (from Steps 10-11) by pipetting with a 200µL pipette set to 30 µl
To each of the four tubes with beads containing the T4 polymerase mix add 6.3µLof splint, using a new pipette tip each time
Place the tubes with resuspended beads into the thermomixer at 37°C and shake for 0h 5m 0s at300rpm
After the incubation at Step 29, place the tubes on a (non-magnetic) rack at Room temperature
Add 6.3µLT4 polymerase to each of the four tubes
Place the tubes on a MacsMix tube rotator for 0h 10m 0sand rotate on second speed setting (at 16rpm)
Transfer the tubes to a thermomixer at 75°C for 0h 10m 0s without shaking
During the 10 minutes incubation time in Step 33, prepare lambda exonuclease mix : in four 1.5 mL DNA LoBind tubes, pipette 95µL lambda exonuclease buffer, 832µL water in each tube and place On ice. Once the incubation at Step 33 is finished, place the tubes On ice for 0h 1m 0s
Wash BD Rhapsody beads as described above in the section Washing BD Rhapsody Beads ( ), after which resuspend in at least 200µL TE/TW buffer and place On ice
Step 4 modification of full vial of RoCKseq beads: lambda exonuclease digest
Place the four tubes containing the beads on a 1.5 mL magnetic stand, wait for 0h 1m 0s and remove the supernatant, not disturbing the beads. The tubes remain on the stand.
Remove the first tube from the stand and resuspend the beads using the lambda exonuclease mix (927µL, from Step 34)
Place the tube on a non-magnetic rack at Room temperature
Repeat Steps 37-38 with other three tubes
To each of the four tubes with beads resuspended in lambda exonuclease mix add 21µLof lambda exonuclease
Transfer the four tubes to a thermomixer at37°Cfor 0h 30m 0s without shaking
Transfer the tubes to a thermomixer set to 75°Cfor 0h 10m 0s without shaking
After the incubation at Step 42, immediately place the tubes On ice for 0h 1m 0s
Wash BD Rhapsody beads as described above in the section Washing BD Rhapsody Beads ( ), after which resuspend in at least 200µL TE/TW buffer and place On ice
Step 5 modification of full vial of RoCKseq beads: final resuspension beads and storage
Place the four tubes containing the BD Rhapsody beads on the 1.5 mL magnetic stand and wait for0h 1m 0s
Remove supernatant from first tube and resuspend the beads in 250µL TE/TW buffer by gently pipetting up and down at least 5 times and place On ice
Repeat step 46 with the other three tubes
Pool the resuspended beads into a new 1.5 mL Lobind tube
Store RoCKseq modified beads at 4°C . Beads are stable over time in TE/TW buffer.
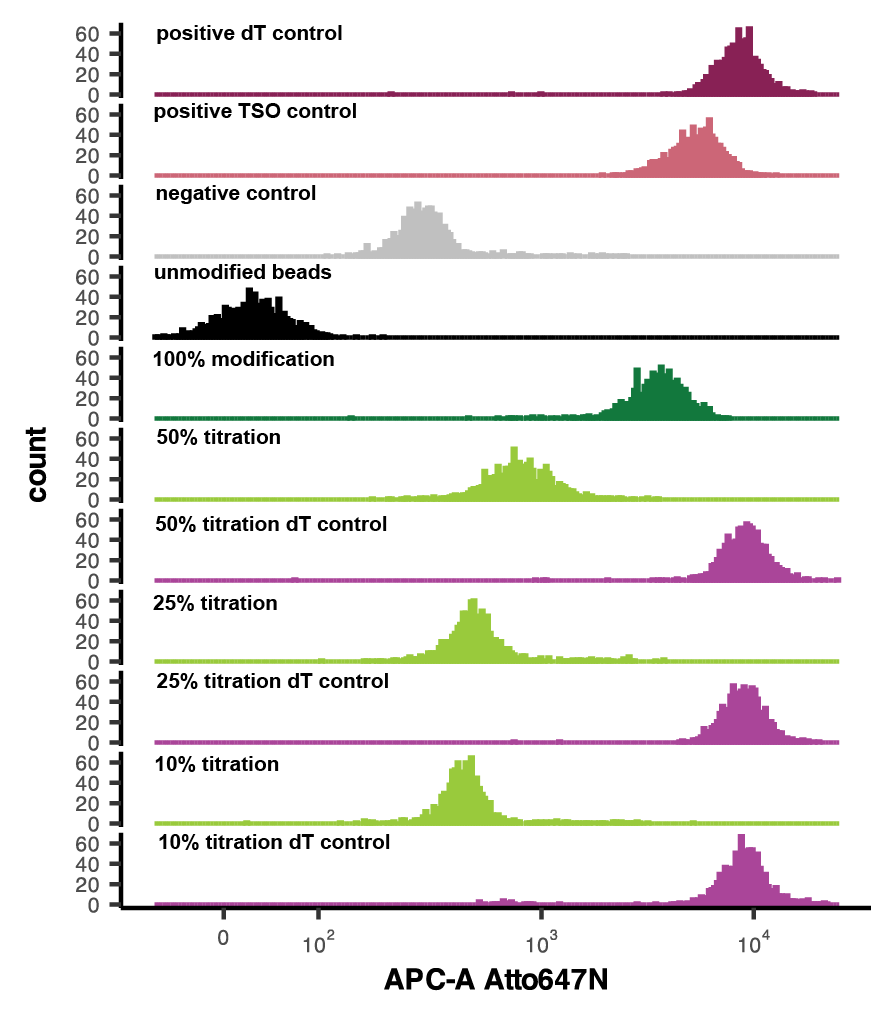
Fluorescent assay for the detection of RoCKseq modification and integrity of DNA oligos on beads
Recommended conditions for fluorecent assay
| A | B | C |
|---|---|---|
| Positive control dT | Barcoded beads (unmod) | polyA fluo oligo |
| Positive control TSO | Barcoded beads (unmod) | TSO fluo oligo |
| Negative control | Barcoded beads (unmod) | Fluo oligo for modification |
| RoCKseq beads | Barcoded beads (modified) | Fluo oligo for modification |
| dT control RoCKseq beads | Barcoded beads (modified) | polyA fluo oligo |
| Unmodified beads | Barcoded beads (unmod) | ----- |
Preheat a thermomixer to 46°C
Prepare TE/TW and Water buffers in 50 mL Falcons and place On ice
Thaw fluorescent oligos (100 µM) at Room temperature
Prepare lysis buffer in a 1.5 mL Eppendorf tube and place On ice: per sample add 1µL1 M DTT (part number 650000063, BD RhapsodyTM Enhanced Cartridge Reagent Kit) to 188µL µl BD Rhapsody lysis buffer (part number 650000064, BD RhapsodyTM Enhanced Cartridge Reagent Kit) and mix thoroughly
Dilute 10µLof fluorescent oligo (100 µM) 1:10 in ddH2O and place On ice. Keep in the dark
Place previously modified BD Rhapsody barcoded beads On ice
Wash unmodified beads used as controls as described in Step 2 modification of full vial of RoCKseq beads: washing BD Rhapsody beads ( ) and place On ice
Pipette 20µLof RoCKseq modified beads per condition in a new Eppendorf tube and place On ice
Place the tubes containing the 20µL of beads on a 1.5 mL magnetic stand, wait for 0h 1m 0s and remove the supernatant, not disturbing the beads. The tubes remain on the stand.
Add 188µL lysis buffer + DTT (from Step 54) per condition and place the tube on a non-magnetic rack at room temperature
Add 8µLof the 10 µM fluorescent oligo per sample (prepared at Step 55) and gently pipette up and down to mix
Incubate samples for 0h 30m 0s at 46°C shaking at 300rpm in the dark (for example covering the thermomixer block with aluminum foil)
Wash beads as described in Step 2 modification of full vial of RoCKseq beads: washing BD Rhapsody beads ( ) and resuspend in 300µL TE/ TW buffer
Strain beads in a Falcon 5 mL Round Bottom Polystyrene Test Tube, place On ice, keep in dark and measure fluorescent intensity at a FACS analyser.
Vortex beads before loading the sample. If the event rate drops, stop acquisition and vortex beads again.
Measure 1000 events per sample.
Design of ROIseq primers
ROIseq primers should be designed directly 5’ (max. 10bp upstream) to the region of interest (ROI). The length of the primers is 12 nucleotides. Since 12 nucleotides will be included in the cDNA sequencing read (HTS), the ROIseq primer must be in close proximity to the ROI.
Depending on the ROI to be detected, it may be advantageous to position the ROIseq primer further upstream to the sequence of interest. This is the case for example for fusion transcripts, in which having a longer stretch to map on both sides of the fusion breakpoint is beneficial. In this case we recommend using longer read length and placing the ROIseq primer 20-30 bp upstream to the ROI itself.
The ROIseq primer has the following structure:
5’-TCAGACGTGTGCTCTTCCGATCTNNNNNNNNNNNN-3’ , the N being the sequence of the ROIseq primer which identical to the coding strand.
An additional consideration when designing ROIseq primers is that ideally the read generated after ROIseq priming should be unique, i.e. doesn’t map to multiple loci.
ROIseq primers should be ordered in HPLC grade and at 0.2 µmol scale and resuspended in DNA Supension buffer from Teknova (T0221).
RoCK and ROI library generation
RoCK and ROI library generation follows the standard BD Rhapsody workflow (mRNA capture, reverse transcription and exonuclease treatment: Doc ID: 210966; library generation Doc ID: 23-21711-00) with the following adaptations (steps 67.1-67.4 indicate the steps in the standard protocols where the changes occur):
Resuspending barcoded beads prior to loading on cartridge: to account for the bead loss during modification, resuspend the RoCKseq beads in 680µLSample Buffer (Cat. No. 650000062, BD RhapsodyTMEnhanced Cartridge Reagent Kit) instead of 750µLprior to loading on the BD Rhapsody cartridge
Random priming and extension : if a single ROIseq primer is added, dilute 1µL of the 100 µM primer 1:10 in ddH2O and pipette 4µLof the diluted mix during the Random Priming and Extension step (after pipetting the 174µL ). Add the ROIseq primers after the beads are resuspended in the Random Primer mix .
If multiple ROIseq primers are used, mix 1µL of each ROIseq primer (100 µM), add ddH2O up to10µLand add 4µL to the mix.
RPE PCR: add 1µL of 100 µM T primer to each sample after the RPE PCR mix is added to the Purified RPE product .
Indexing PCR : for indexing of RoCKseq libraries, a separate PCR is performed substituting 5µL of the Library Forward Primer (BD RhapsodyTMEnhanced Cartridge Reagent Kit, part number 91-1085) with 5µL of 100 µM of a custom indexing primer. The same primary library and reverse primers are used as recommended by the manufacturer. The reaction is thus as follows:
For WTA library (from BD Rhapsody Doc ID: 23-21711-00):
| A | B | C | D |
|---|---|---|---|
| PCR MasterMix (Cat. No. 91-1118) | 25 | 30 | 55 |
| Library Forward Primer (Cat. No. 91-1085) | 5 | 6 | 11 |
| Library Reverse Primer (1-4)(Cat. Nos. 650000080, 650000091-93) | 5 | 6 | – |
| Nuclease-free water (Cat. No. 650000076) | 5 | 6 | 11 |
| Total | 40 | 48 | 77 |
For TSO library:
| A | B | C | D |
|---|---|---|---|
| PCR MasterMix (Cat. No. 91-1118) | 25 | 30 | 55 |
| T primer + adapter | 5 | 6 | 11 |
| Library Reverse Primer (1-4)(Cat. Nos. 650000080, 650000091-93) | 5 | 6 | – |
| Nuclease-free water (Cat. No. 650000076) | 5 | 6 | 11 |
| Total | 40 | 48 | 77 |
IMPORTANT: RoCKseq and dT-based libraries of a given sample should be indexed with the SAME BD Rhapsody Library Reverse Primer and will thus have the same 8 bp index. The two data modalities are then separated bioinformatically (see Step 69)
If no ROIseq is being performed omit step 67.2
Sequencing
We recommend pooling the WTA and TSO libraries in a 1:1 ratio.
For sequencing of pooled libraries including at least one RoCKseq modified sample (with or without ROIseq primers), a custom R1 primer should be spiked in (see Materials).
The length of R1 should be 60 bp, while the length of R2 may vary depending on the ROI of interest (see section Design of ROIseq primers, Step 66). We recommend using an R2 of 62 bp for ROIs such as point mutations and splice junctions and an R2 of 150 bp for fusion breakpoints and CRISPR target sites.
Data analysis
RoCK and ROI data can be analysed using our custom pipeline, found at https://zenodo.org/records/11070201 under the GPLv3 terms. For downstream data processing please see https://zenodo.org/records/11124929. https://zenodo.org/records/11124929.