Recombinant protein expression and purification of Taq DNA polymerase
Maira Rivera, Javiera Reyes, Javiera Avilés, Amparo Núñez, Fernan Federici, Cesar A Ramirez-Sarmiento
Abstract
This is a slightly modified and simplified version of a protocol by Thomas G.W. Graham et al, which is available at https://gitlab.com/tjian-darzacq-lab/bearmix and has been described in depth in the article 10.1371/journal.pone.0246647, for the recombinant expression of a E602D mutant of Taq DNA polymerase in pET-28a that is available in Addgene (Addgene plasmid # 166944 ; http://n2t.net/addgene:166944 ; RRID:Addgene_166944).
The main goal of this protocol is to eliminate the use of large volumes for dialysis and potential issues with the protein crashing out of the solution due to the use of concentrators for buffer exchange of this enzyme into storage conditions.
Steps
DAY 1 – Plasmid transformation
Transform 100ngof plasmid containing Taq DNA polymerase into E. coli C41 competent cells using either heat shock or electroporation.
Spread transformed cells in LB Agar plates supplemented with 0.05mg/mLKan. Grow plate overnight at 37°C.
DAY 2 – Preinoculum
Select a single colony from the LB agar plate to prepare a preinoculum in 10mL LB media supplemented with 0.05mg/mLKan. Grow overnight at 200rpm.
DAY 3 – Protein Overexpression
Use the full volume of the preinoculum to inoculate 1L of LB (or TB) media supplemented with 0.05mg/mLKan (1% inoculation). Grow at 160rpm until reaching an optical density at 600 nm (OD600) = 0.8.
Upon reaching OD600= 0.8, add IPTG to a final concentration of 0.5millimolar (mM) and incubate 160rpm.
DAY 4A – Protein Purification by IMAC
Centrifuge the cell culture 4000x g,4°C.Then, resuspend the cell pellet in 50mL of Buffer A freshly supplemented with 1.0millimolar (mM) PMSF and 0.2mg/mL lysozyme.
Incubate the resuspended cells at 80rpm.
Sonicate on ice for 0h 8m 0s using cycles of 0h 0m 1s ON and 0h 0m 6s OFF at 40% amplitude (Qsonica Q125, 125W).
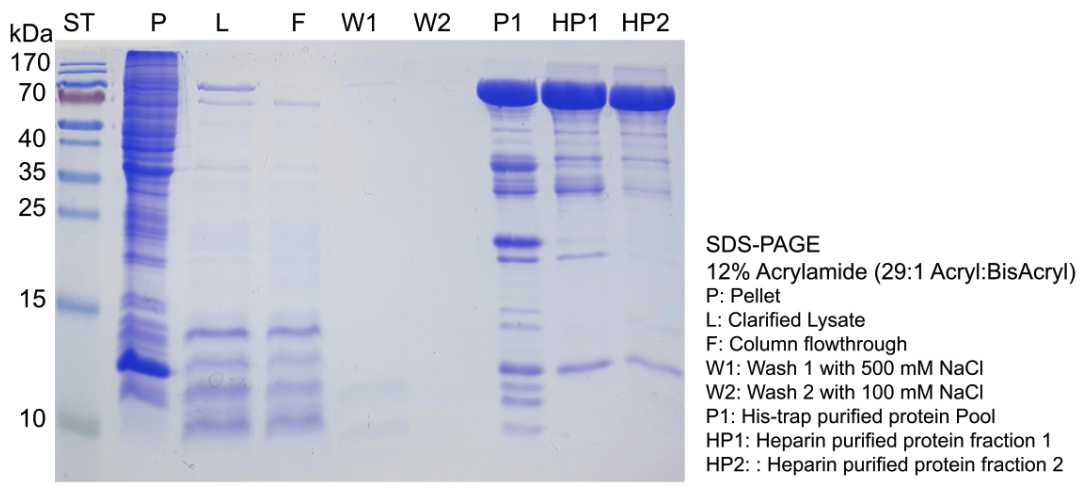
On an ultracentrifugation tube, incubate the unclarified lysate at 75°C for 0h 30m 0s to precipitate most of E. coli proteins, and then place on ice for 0h 5m 0s. Centrifuge 20000x g,4°C and collect the supernatant. You might want to collect a small sample for SDS-PAGE afterwards.
On a 1 mL HisTrap column (GE Healthcare) pre-equilibrated with 10 column volumes (c.v.) (here, 10 mL) of Buffer A , load the supernant. Wash with 10-20 c.v. of Buffer B . Repeat the wash step with Buffer C . Then, elute with 5 c.v. of Buffer D , collecting the eluted fractions every 0.5mLin 1.5 ml tubes.
To quickly pool the fractions containing the protein of interest, prepare a 96-well plate or 1.5 mL tubes with 40µLof 5X Bradford reagent and 150µLof distilled water. Then, add 10µLof each protein fraction and compare against a blank reference sample corresponding to 10µLof Buffer C . You can determine your protein-containing fractions either by absorbance at 595 nm on a plate reader or visually by comparing the blue coloration of each fraction against the blank reference. Pool your fractions and collect a 10µL sample for SDS-PAGE.
DAY 4B – Second purification and buffer exchange by Heparin
This method was preferred over protein dyalisis or Amicon protein concentration to avoid using large buffer volumes and proteins crashing out of the solution.
Dilute the pooled fractions 6X in buffer containing 50 mM Tris-HCl pH 8.0 and 100 mM NaCl.
Next, load the diluted sample onto a 1 ml HiTrap Heparin column previously equilibrated with 10 c.v. (here, 10 mL) Buffer HA . Then, elute the protein using a 10 c.v. linear gradient against Buffer HB , collecting the eluted fractions every 0.5mLin 1.5 ml tubes.
This linear gradient can be achieved by connecting two containers, one with 5 c.v. Buffer HA and the other with 5 c.v. buffer HB, with a syphon or a tube, and withdrawing solution from the Buffer HA container to the column using a cheap peristaltic pump or by gravity.
Again, determine your protein-containing fractions using the Bradford assay. Pool your fractions and determine its protein concentration using the same method and collect a 10µL sample for SDS-PAGE.
For storage, supplement your pooled fractions with 100millimolar (mM) Tris-HCl pH 8.0, 0.2millimolar (mM) EDTA and 6millimolar (mM) DTT. Then, dilute the sample by adding glycerol up to 50% volume to reach final storage conditions: 50 mM Tris-HCl pH 8.0, ~100 mM NaCl, 0.1 mM EDTA, 3 mM DTT.
Generate 200µL aliquots of the enzyme and store it at -20°C until required.