RSV whole genome sequencing using ONT
Kseniia Komissarova
Abstract
Respiratory Syncytial Virus (RSV) is a leading cause of hospitalization due to acute lower respiratory infection in infants and young children. Monoclonal antibodies palivizumab is available option for treating RSV limited to select populations in high-resource settings. There is no vaccine to prevent RSV to the moment. Fortunately, several vaccine candidates are now in the human testing phase targeting young children, older adults and pregnant women, and an effective safe vaccine is likely to be available in the near future.
For vaccine and therapy development RSV surveillance is essential. Using whole-genome sequencing data helps ensure the vaccine has the most up-to-date coverage and more-effective global reach. Through genomic surveillance we can track the spread of variants, monitor changes to the genetic code of the virus. Finally, this data is used to better understand how virus might impact public health.
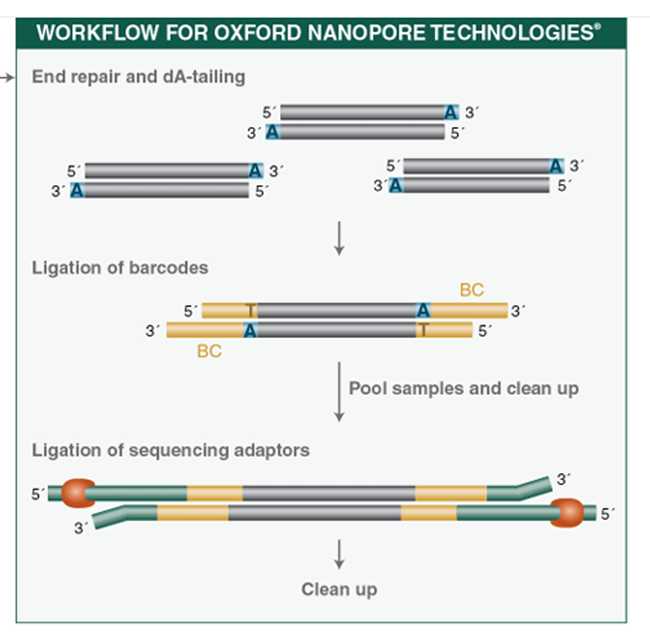
Here we provide the protocol for genome sequencing of RSV A using Oxford Nanopore Technology platform. The four overlapping genome fragments together comprise of the full RSV genome encompassing all viral genes, yet lacking the far 3′ and 5′ genome termini.
Steps
Order primers according to the list from your local manufacturer
| A | B |
|---|---|
| RSVA-fragment 1-Fw | AAAAATGCGTACWACAAACTTGC |
| RSVA-fragment 1-Rev | GTTGGTCCTTGGTTTTGGAC |
| RSVA-fragment 2-Fw | CACAGTGACTGACAACAAAGGAG |
| RSVA-fragment 2-Rev | GCTCATGGCAACACATGC |
| RSVA-fragment 3-Fw | CGAGGTCATTGCTTGAATGG |
| RSVA-fragment 3-Rev | CACCACCACCAAATAACATGG |
| RSVA-fragment 4-Fw | AGGGTGGTGTCAAAAACTATGG |
| RSVA-fragment 4-Rev | ACGAGAAAAAAAGTGTCAAAAACT |
Primer sequences designed by Langedijk, A.C., Lebbink, R.J., Naaktgeboren, C.et al. Global molecular diversity of RSV – the “INFORM RSV” study.BMC Infect Dis 20, 450 (2020). https://doi.org/10.1186/s12879-020-05175-4
Dilute primers to the concentration of 10mM. Aliquote and freese.
Briefly vortex and centrifuge defrosted reagents before use.
Prepare four RT-qPCR reactions for every RNA sample you need to amplify for sequencing.
Use one pair of primers above (forward and reverse) per reaction
Prepare the PCR reaction mixture following the specifications below:
| A | B |
|---|---|
| primer F (10μM) | 0,5 |
| primer R (10μM) | 0,5 |
| SuperScript III RT/Platinum Taq Hi-Fidelity Enzyme Mix | 0,5 |
| 2x reaction mix | 12,5 |
| template RNA | 5 |
| water | 6 |
SuperScript™ III One-Step RT-PCR System with Platinum™ Taq High Fidelity DNA Polymerase is recommended for RT-PCR
Use next cycling conditions for your thermocycler:
-
55 °C for 10 min
-
98 °C for 2 min,
-
40 cycles of 98 °C for 10 s, 61 °C for 10 s and 72 °C for 3 min
Assess quantity and quality of amplicons using capillary or gel DNA electrophoresis.
Lenght of the products are: 4665, 4086, 3802, 4431 b.p.
For each sample combine four PCR reactions in one volume (100 mcl summary)