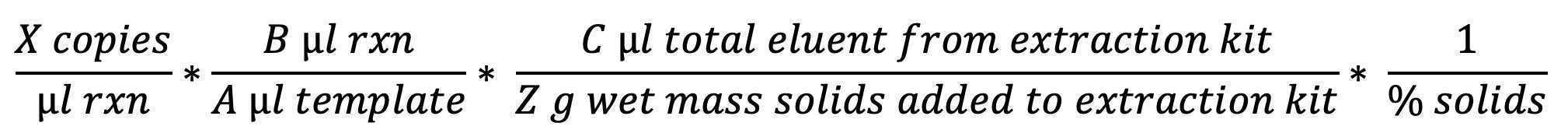Quantification of various SARS-CoV-2 variant mutations (characteristic of Alpha, Beta, Gamma, Delta, Omicron and Omicron sublineages) in settled solids using digital RT-PCR
Alexandria B Boehm, Bridgette Hughes, Bradley J. White, Marlene K. Wolfe
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
This process instruction describes the steps for quantitative analysis of nucleic acid from SARS-CoV-2 with a triplex Reverse Transcriptase droplet digital Polymerase Chain Reaction (RT-ddPCR) assay targeting the N Gene, S Gene and various mutation assays in extracted and purified RNA samples from solid wastewater samples for population level SARS-CoV-2 community surveillance. RT-ddPCR is a modified version of conventional RT-PCR workflows which involves separating the reaction mixture into many partitions (~20,000) before thermal cycling which allows for direct absolute quantification of the target RNA molecules.
Future protocols will be published that are complementary to this one and describe assays targeting additional SARS-CoV-2 mutations.
This protocol uses RNA extracted using this protocol: High Throughput RNA Extraction and PCR Inhibitor Removal of Settled Solids for Wastewater Surveillance of SARS-CoV-2 RNA. That RNA is generated from samples subjected to pre-analytical steps outlined in: High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses. It is recommended that these assays be run along assays for PMMoV and BCoV as controls as described in the companion protocol High Throughput SARS-COV-2, PMMOV, and BCoV quantification in settled solids using digital RT-PCR
The readout of this assay is a concentration of each target in the extracted RNA samples (copies/µL).
Scope
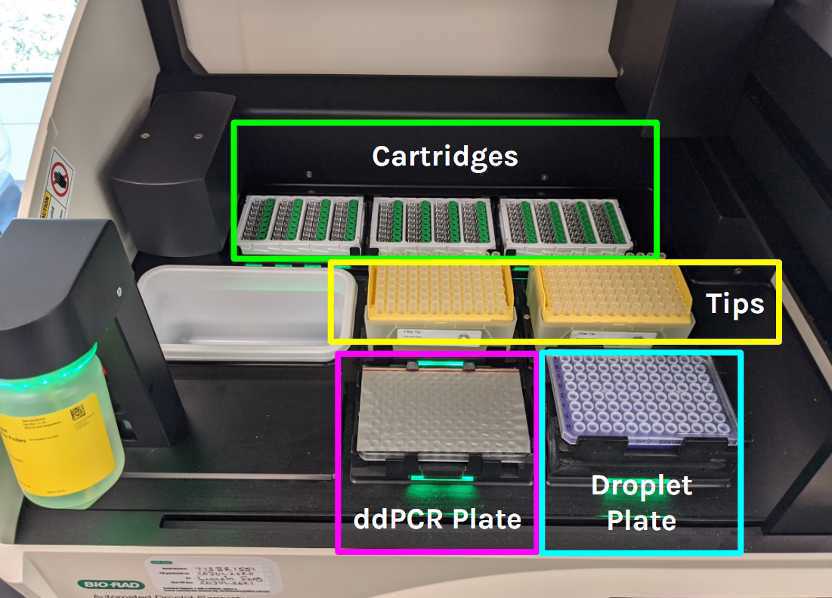
This process instruction applies to quantitative analysis of nucleic acid from SARS-CoV-2 RNA from solid wastewater samples with ddPCR using a Bio-Rad AutoDG Droplet Digital PCR system consisting of the AutoDG Automated Droplet Generator and the QX200 droplet reader.
Publications
The following publications describe use of these assays:
M. K. Wolfe, B. Hughes, D. Duong, V. Chan-Herur, K. R. Wigginton, B. White, A. B. Boehm. 2022. Detection of SARS-CoV-2 variant Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in wastewater settled solids using mutation-specific assays is associated with regional detection of variants in clinical samples. Applied and Environmental Microbiology . 88(8), e00045-22.Link.
A. B. Boehm, B. Hughes, M. K. Wolfe, B. J. White, D. Duong, V. Chan-Herur. Regional replacement of SARS-CoV-2 variant Omicron BA.1 with BA.2 as observed through wastewater surveillance. Accepted to Environmental Science & Technology Letters .Link to preprint.
Kirby AE, Welsh RM, Marsh ZA, Yu AT, Vugia DJ, Boehm AB, Wolfe MK, White BJ, Matzinger SR, Wheeler A, Bankers L, Andresen K, Salatas C, NYDEP, Gregory DA, Johnson MC, Trujillo M, Kannoly S, Smyth DS, Dennehy JJ, Sapoval N, Ensor K, Treangen T, Stadler LB, Hopkins L. 2022. Notes from the Field: Early Evidence of the SARS-CoV-2 B.1.1.529 (Omicron) Variant in Community Wastewater - United States, November - December 2021. MMWR Morb Mortal Wkly Rep 2022;71:103- 105. DOI:http://dx.doi.org/10.15585/mmwr.mm7103a5.
A. Yu, B. Hughes, M. Wolfe, T. Leon, D. Duong, A. Rabe, L. Kennedy, S. Ravuri, B. White, K. Wigginton, A. Boehm, D. Vugia. 2022. Estimating relative abundance of two SARS-CoV-2 variants through wastewater surveillance at two large metropolitan sites. Emerging Infectious Disease , 28(5), 940-947.Link.
Steps
Preparation
Retrieve all kit components from the One-Step RT-ddPCR advanced kit for probes from the -20°C freezer and thaw the components -20On ice.
Retrieve ddPCR positive control aliquots (50 copies per uL gRNA) from the -80°C freezer and thaw -20On ice
For re-running frozen plates only:
Thaw the RNA storage plate -20On ice. Before analysis of thawed frozen samples, ensure that the plate is adequately sealed and briefly vortex and centrifuge the plate to ensure the samples are well mixed.
Keep extracted RNA samples -20On ice or in a cold block from the freezer at all times.
ddPCR master mix preparation
Prepare Master Mix
Prepare a working stock of fresh nuclease free water by pouring from a 50 mL Ambion Nuclease Free water into a 5mL eppendorf tube.
Store Master Mix components -20On ice as much as possible during the preparation process.
Briefly vortex One-Step RT supermix for Probes (Yellow Tube), Reverse Transcriptase (Orange Tube), and DTT (Grey Tube) to mix contents then briefly spin with the benchtop centrifuge.
In a 2mL eppendorf tube, prepare the master mix according to the table for SARS-CoV-2 quantification with HV69-70, E484K/N501Y, or del156-157/R158G. Store prepared master mix on ice.
Primer/Probes Mixes
| A | B | C |
|---|---|---|
| 20x Concentration | Final Concentration/Rxn | |
| Primers (each) | 18 µM | 900 nM |
| N Probe (FAM) | 5 µM | 250 nM |
| S Probe (HEX) | 5 µM | 250 nM |
| XXX Probe (HEX) | 2.5 µM | 125 nM |
| XXX Probe (FAM) | 2.5 µM | 125 nM |
The 20x SARS-CoV-2 and XXX Primer/Probe Mix contains primers and probes in the following concentrations (suspended in molecular grade water). XXX is the mutation of interest.
Master Mix
| A | B | C |
|---|---|---|
| Reagents | Volume per Well | Volume Per Plate |
| ddPCR™ One-Step RT supermix for Probes (Yellow Tube) | 5.5 µL | 580.8 µL |
| 20x SARS-CoV-2 Primer/Probe Mix | 3.3 µL | 348.48 µL |
| Reverse Transcriptase (Orange Tube) | 2.2 µL | 232.32 µL |
| 300mM DTT (Gray Tube) | 1.1 µL | 116.16 µL |
| Nuclease Free Water | 4.4 µL | 464.64 µL |
| Total Volume | 16.5 µL | 1742.4 µL |
SARS-CoV-2 ddPCR Master Mix. Volume per plate assumes 96 well plate (with 10% excess).
Transfer Master Mix and Samples to ddPCR plate
For each assay, remove a cold block from the freezer and place a new Bio-Rad ddPCR 96-well Plate in it.
Using a new reagent reservoir, manually pipette 16.5µL of the appropriate Master Mix to each well of each ddPCR Plate.
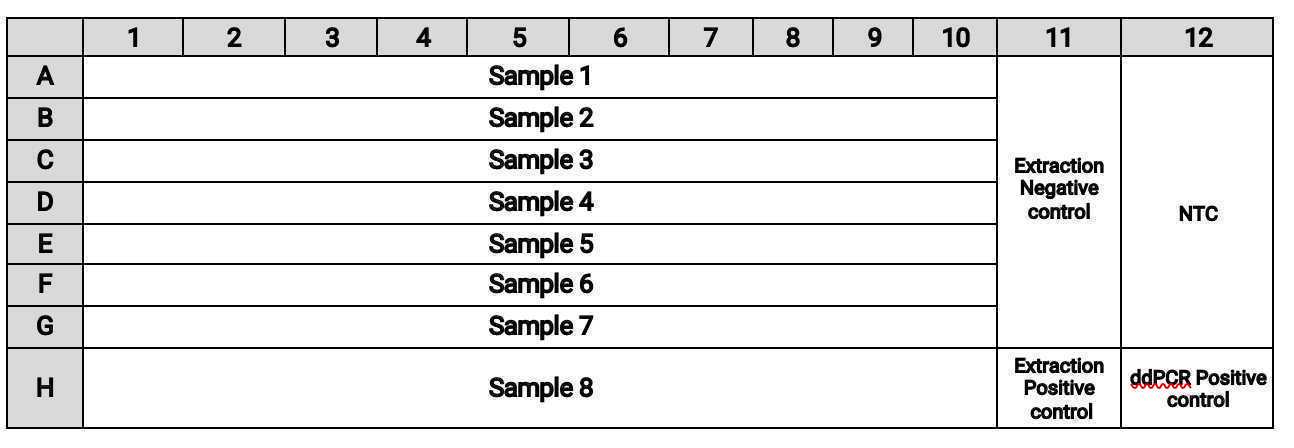
Transfer samples to the following wells on the plate either manually or using a liquid handling robot such as the Agilent Bravo system:
- Transfer
5.5µLof samples from columns 1-10 of the RNA plate into columns 1-10 of the ddPCR plate. - Transfer
5.5µLof the extraction controls from column 12 of the RNA plate into column 11 of the ddPCR plate. - Transfer
5.5µLof NTC (water) into A-G of column 12. - Add
5.5µLof ddPCR positive control to well H12 of the ddPCR plate (add manually even if using a liquid handler)NoteUse SARS-CoV-2 genomic RNA (ATCC® VR-1986D™) mixed 1:1 with appropriate mutation standard as ddPCR positive control (see materials section of the protocol). The extraction positive controls are generated during RNA extraction per this companion protocol: Use SARS-CoV-2 genomic RNA (ATCC® VR-1986D™) mixed 1:1 with appropriate mutation standard as ddPCR positive control (see materials section of the protocol). The extraction positive controls are generated during RNA extraction per this companion protocol: https://www.protocols.io/view/high-throughput-rna-extraction-and-pcr-inhibitor-r-btyrnpv6. In that protocol, the extraction positive control consists of SARS-CoV-2 genomic RNA that does not contain the mutations HV69-70 and E484K/N501Y or other mutation targets. . In that protocol, the extraction positive control consists of SARS-CoV-2 genomic RNA that does not contain the mutations HV69-70 and E484K/N501Y or other mutation targets.
The plate layout should be:
Bring the BioRad ddPCR plate with Master Mix and samples to the Bio-Rad PX1 PCR Plate Sealer and place it in the metal carrier inside the plate sealer.
Align a PCR Plate Heat Seal Foil on top of the ddPCR plate with the red line facing up.
Seal the plate by pressing the green “seal” button.
Briefly vortex the plate using a bench top plate vortexer.
Briefly spin down the plate in the bench top Axygen Plate Centrifuge.
Store the ddPCR plate on a white cold block or On ice until droplet generation.
ddPCR droplet generation
Generate Droplets:
On the AutoDG droplet generator: select “configure plate” and then press the blue arrow in the upper left corner to highlight all columns.
Load 3 DG32 cartridge plates and 2 tip boxes with the lid removed onto the AutoDG deck. The icons on the AutoDG display will change from yellow to green if the plates and tips are oriented correctly.
Place the sealed, vortexed, and centrifuged ddPCR plate in the Sample Plate position on the AutoDG deck.
Remove the AutoDG cooling block from the freezer and place it in the Droplet Plate slot on the AutoDG deck.
For the Droplet Plate: label a new Bio-Rad ddPCR™ 96-Well Plate and place it in the Auto DG cooling block.
When droplet generation is done, open the lid and remove the droplet plate from the Auto DG cold block and place it in the metal carrier inside the plate sealer.
Align a PCR Plate Heat Seal Foil on top of the droplet plate with the red line facing up.
Seal the plate by pressing the green “seal” button.
Thermocycling
Place the plate in the thermocycler. Verify and run the thermocycler program according to the table for SARS-CoV-2 ddPCR.
| A | B | C | D |
|---|---|---|---|
| Steps | Cycle #s | Temp (°C) | Time |
| 1 | 1 | 50 | 60 min |
| 2 | 1 | 95 | 5 min |
| 3 | 40 | 95 | 30 secs |
| 40 | 59 | 1 min | |
| 4 | 1 | 98 | 10 min |
| 5 | - | 4 | hold |
Cycling conditions for the N/S/HV69-70 multiplexed assay
| A | B | C | D |
|---|---|---|---|
| Steps | Cycle #s | Temp (°C) | Time |
| 1 | 1 | 50 | 60 min |
| 2 | 1 | 95 | 5 min |
| 3 | 40 | 95 | 30 secs |
| 40 | 61 | 1 min | |
| 4 | 1 | 98 | 10 min |
| 5 | - | 4 | hold |
Cycling conditions for the N/S/XXX multiplexed assay where XXX are all other mutations.
After the PCR program is done, store the ddPCR plate at 4°C (in the thermal cycler, on ice or in the fridge) until droplet reading.
Read Droplets
Before running the plate on the QX200 droplet reader, open the drawer on the left side of the instrument and check that there is adequate droplet reader oil and that the waste bottle is not overly full.
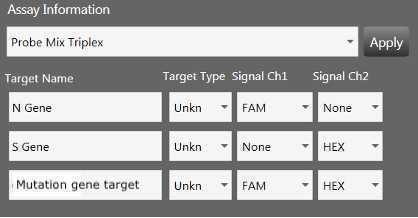
Load template from previous plate of the same assay - ensure that all parameters (supermix, dye, absorbance etc.) are correct for the assay used and that sample names are assigned to the appropriate wells. Save as with a new plate name.
Name the file with a name clearly describing which plate you are reading.
Remove the plate from the thermal cycler and secure it in the droplet reader by placing it in the stage, placing the metal brace on top of it and pressing down the black plastic tabs on either side of the brace.
Click “Run” in Quantasoft to commence droplet reading.
Select “Ok” when the next dialog box comes up. Do not change the settings from “Columns” and “FAM/HEX”.
Post-Processing Analysis
SARS-CoV-2 triplex assay analysis
Open QuantaSoft™ Analysis Pro Software
Click on the Table Menu Button on the right side of the Well Data table and select Export to CSV to export data.
Save the QuantaSoft Analysis
Open the .qlp file associated with the run
At this point, if sample names were not added at the start of the run, fill in the sample name by selecting the wells, typing in sample ID and click “Apply”.
If wells need to be deselected: Go to the Plate Editor tab, select wells to exclude and click “Clear Selected Well”.
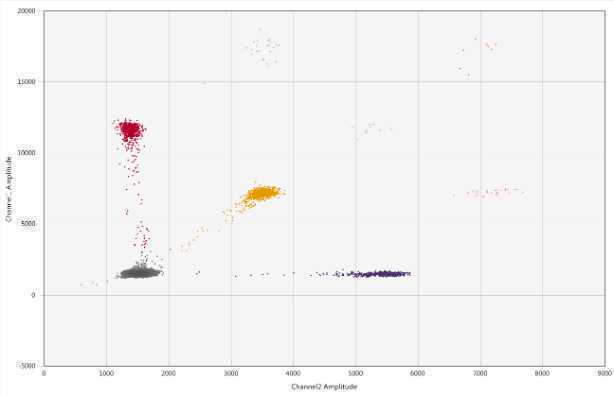
Go through every well to ensure that:
- The droplets are designated properly per well
- There are >10,000 droplets per well
If any well does not have the appropriate droplets, go to the Plate Editor tab, select the affected well and click “Clear Selected Well”.
On the 2D Amplitude tab, select Merged Wells on the left side of the page
Dimensional Analysis and Quality Control
For dimensional analysis to express the results of each assay in terms of gene copies/dry weight solids:
Begin with the concentration provided by the QuantaSoft software and reported in the CSV, as expressed in gc/uL of reaction.
To determine the concentration in cp/g dry weight: