Quantification of tissue creatine content using capillary electrophoresis
Jessica Terrill, Jane Choi, Angelo Bautista, J. Jane Pillow, Peter G Arthur
Disclaimer
No competing interests are declared.
Abstract
The current protocol describes an alternative method for creatine quantification in biological tissue samples using capillary electrophoresis, with high separation efficiency that enables rapid analysis (<10 minutes) with low sample volumes. The protocol involves homogenization of snap-frozen tissue samples in phosphate buffer, followed by electrophoresis through a bare-fused capillary (75 µm i.d.) and measurement at 200 nm. Under the optimised conditions, we found excellent linearity in creatine standards between 6.3 – 100 µM. The overall intra-assay variability for concentrations between 6.3 – 100 µM was 1.5 %, and the inter-assay variability was 6.4 %, with a limit of detection at 6 nmol/mg protein. Variables such as the amount of sample injected or the capillary diameter could be changed to enhance the lower limit of detection. The current protocol was developed and optimised using ovine lung tissue samples, but it can be easily adapted to analyse various tissue types.
Steps
Preparation of buffers
Prepare the stock phosphate buffer (PB) 200millimolar (mM) 6
Add 1199.8 mg NaH2PO4 to 30 mL ddH2O, dissolve well
pH the solution to 6.0 with concentrated NaOH
top up the volume to 50 mL with ddH2O (ideally using a volumetric flask)
filter sterilise the solution through a 0.45 µm syringe filter
store at 4°C for a maximum of 4 weeks. Discard if any precipitate forms.
M × V × #H2O × 18.015 = mL H2O contributed by the hydrated compound.
Prepare the background electrolyte (BGE) 75millimolar (mM) 150millimolar (mM) 6
Dilute 18.75 mL of 200 mM PB buffer (pH = 6) with 20 mL ddH2O
Add 2.163 g SDS to the solution and dissolve well
Top up the volume to 50 mL with ddH2O using a volumetric flask
Filter sterilise the solution through a 0.45 µm syringe filter
Store at Room temperature for up to a week. Discard if any precipitate forms.
Prepare the homogenisation buffer (S-BGE) 50millimolar (mM) 150millimolar (mM) 0.01% volume 6
Dilute 12.5 mL of 200 mM PB buffer (pH=6) with 17.5 mL ddH2O
Add 2.163 g SDS to the solution and dissolve well
Top up the volume to 50 mL with ddH2O using a volumetric flask
Remove 5 µL of solution and replace with 5 µL DMSO
Filter sterilise the solution through a 0.45 µm syringe filter
Store at Room temperature for up to a week. Discard if any precipitate forms.
Preparation of creatine standard
Prepare creatine standards by dissolving creatine monohydrate (C3630, Merck) or anhydrous (C0780, Merck) to a stock concentration of 10 mM.
Note : If using creatine monohydrate – account for the hydrated compound by excluding the mass
of 22O from calculations(Adams, 2008)). Although this is negligible e at small volumes/concentrations.
M x V x #H2O x 18.015 = mL H2O contributed by the hydrated compound.
Dilute the 10 mM stock solution to the appropriate range using the same solvent as the samples . i.e., If running enzymatic analysis – dilute the standards in PBS. If running samples – dilute the standards in the S-BGE (50 mM PB buffer).
Note: Prepare creatine standards within the range typically expected for the tissue type. However, this can also depend on the protein concentration of the samples.
For lung tissue, standard curve concentrations between the 6.25 - 100 µM range have shown excellent linearity with low inter-assay variability.
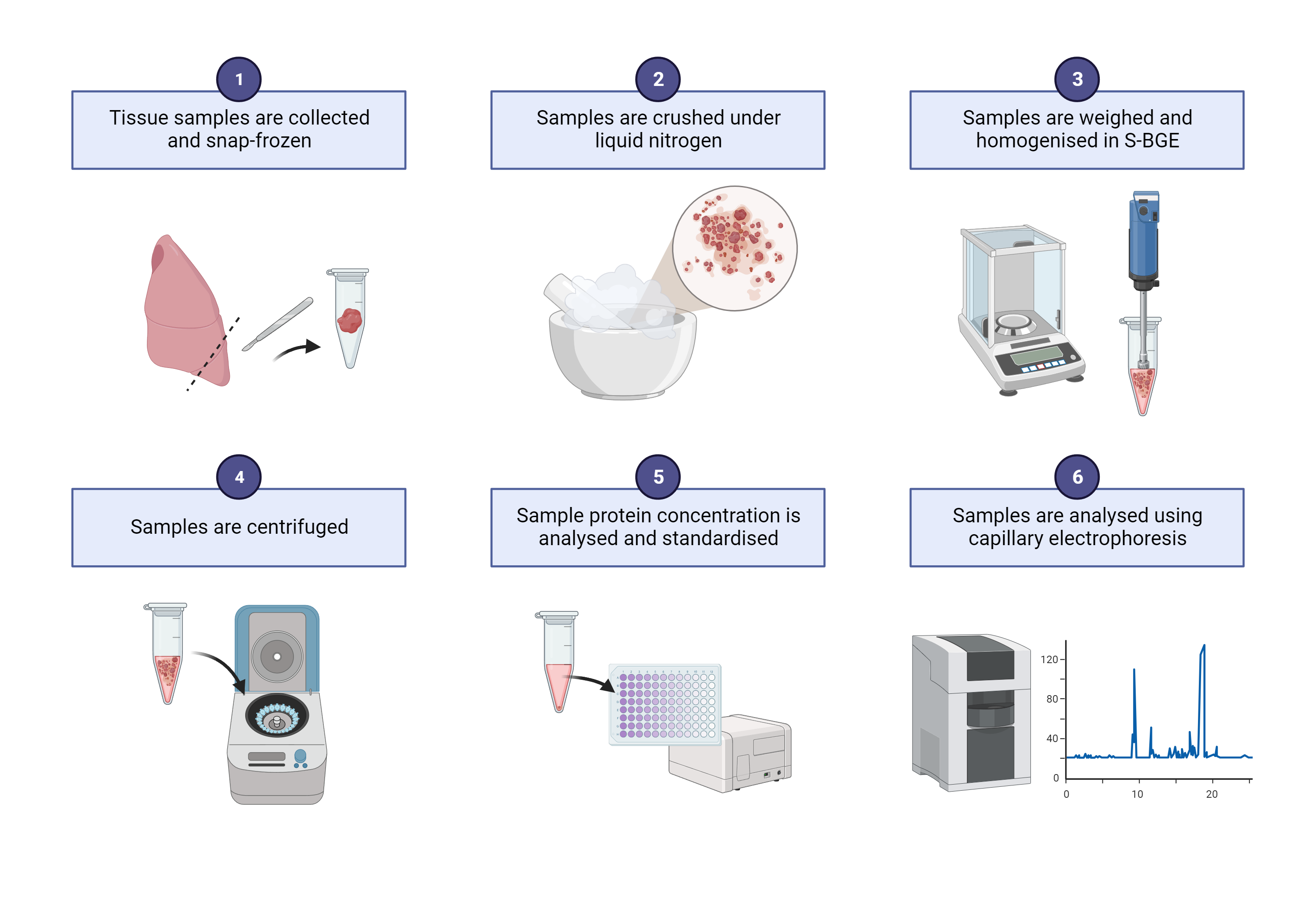
Sample preparation for enzymatic analysis
This step is required once for each sample type to determine and confirm which peak on the electropherogram corresponds to creatine. Validation is performed by adding creatinase enzyme (which breaks down creatine into urea & sarcosine) to deplete the signal. Comparison can be made between samples with and without creatinine to identify the modified peak as creatine.
In essence, the addition of the creatinase enzyme will essentially serve as a negative control.
Rapidly homogenise snap-frozen tissue samples in 20 volume of 1X PBS
a. With a mortar & pestle with liquid nitrogen
b. Or a bead beater
Centrifuge the homogenates 10000x g,4°C
Transfer supernatants into a new tube.
Take an aliquot of the supernatant and determine the protein concentration with an appropriate protein assay (e.g. BCA, Bradford, DC assay)
Dilute the samples with PBS to standardise the protein concentration in all samples (typically 2 mg/mL for lungs).
Aliquot and store at -80 °C until use.
Sample preparation for CE analysis
Freshly prepared S-BGE is recommended for sample homogenisation
Rapidly homogenise snap-frozen tissue samples in 20 volume of S-BGE
a. With a mortar & pestle with liquid nitrogen
b. Or a bead beater
Centrifuge the homogenates 10000x g,24Room temperature
Note, keep samples in RT as SDS will precipitate at 4 degrees.
Transfer supernatants into a new tube
Take an aliquot of the supernatant and determine the protein concentration with an appropriate protein assay (e.g. BCA, DC assay are compatible with SDS in the buffer)
Dilute the samples with S-BGE to standardise the protein concentration in all samples (typically 2 mg/mL for lungs).
Aliquot and store at -80 °C until use.
Equipment setup
Capillary electrophoresis is performed on the Agilent 7100 CE system (Agilent Technologies, CA, USA) with:
- bare-fused silica capillary (75 µm inner diameter; Polymicro Technologies, AZ, USA)
- effective length 36.5 cm
- length to detector 8.5 cm Note: wrap the capillary around the cassette so there is no overlap or touching, as this can generate and spread heat.
Create a new protocol with the separation parameters as follows:
- Injection – 10 mbar for 10 s
- Separation is achieved with a constant voltage of 15 kV applied for 6 mins
- Capillary temperature set at 25 °C
- UV absorbance is measured at 200 nm
Post-conditioning parameters (after each sample) are:
- ddH2O for 30 s
- 0.1 M NaOH for 30 s
- ddH2O for 30 s
- BGE for 120 s
Running the Assay
At the beginning of each run, pre-condition the capillary by flushing in the following order:
- ddH2O for 180 s
- 0.1 M NaOH for 500 s
- ddH2O for 120 s
- BGE for 600 s
Load each tube with at least 10 µL (preferably 20 µL) of sample. Avoid creating bubbles as this can interfere with sample injection. Load the tubes on the Agilent machine.
Run the protocol. The samples can be submitted individually or as a sequence. If running more than ten samples, insert a flush protocol (ddH2O or BGE for 120 s) every ten runs.
Depending on the number of samples to analyse, it may be best to prepare two protocols with two different BGE & waste tubes assigned to ensure sufficient background buffer for the analysis and that the waste doesn't overflow.
At the end of the day, ensure post-conditioning has been successfully run and turn off the equipment.