Quantification of the SARS-CoV-2 using Nanotrap Particles®, the QIAcuity™ Digital PCR System, and GT-Digital SARS-CoV-2 Wastewater Surveillance Assay
Solana Narum, Thibault Stalder, Erik Coats, Eva Top
Disclaimer
FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer-reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
This protocol was developed in an effort to serve as a timely and efficient method for the surveillance of the SARS-CoV-2 in primary influent wastewater samples. This process describes viral concentration via Nanotrap particles, RNA extraction using the Qiagen AllPrep PowerViral DNA/RNA kits with the Qiacube Connect, and quantification of the N1 and N2 genes in SARS-CoV-2 using the GT Digital SARS-CoV-2 Wastewater Surveillance for QIAcuity.
To compile the entire process from beginning to the end some sections were taken from the AllPrep PowerViral DNA/RNA Kit (Qiagen) and the GT Digital SARSCoV- 2 Wastewater Surveillance for QIAcuity v1.0 (GT Molecular) handbooks as well as the Ceres Nano Manual Nanotrap Wastewater Protocol using Qiagen AllPrep PowerViral DNA/RNA Kit protocol (Ceres Nano).
Steps
Sample Collection
Composite primary influent samples are collected (grab samples, flow composite, or time composite)
50mL sub-samples are collected in 50 mL sterile conical tubes. The samples are stored at 4°C until further processing.
Concentration of Viral Fraction
Thaw an aliquot of BCoVworking (-80On ice (see section Preparation of Bovilis Coronavirus). Pipette up and down to mix the aliquot and briefly centrifuge.
Add 62µL of BCoVworking to each 50mL wastewater sample and invert to mix.
Using a serological pipette transfer 10mL of the wastewater sample into a 15 mL conical tube. Repeat this for a duplicate.
Add 100µL of Nanotrap Enhancement Reagent 2 to each 10 mL wastewater sample and vortex for a few seconds.
Add 150µL of Nanotrap Magnetic Virus Particles and invert 2-3 times to mix.
Incubate the samples for 10 minutes at room temperature.
Place the sample tubes in a 15 mL magnetic rack. Wait 1 minute. Remove the supernatant using a serological pipette. The magnetic particles form a pellet on the sides of the tube so it is essential to hold the tip of the pipette in the center of the tube while removing the supernatant. Transfer the supernatant to a waste container.
Add 1mL molecular grade water to each 15 mL tube and remove from magnetic rack. Invert the sample to collect the Nanotrap Magnetic Particles in the water.
Transfer the sample using a pipet to a 1.5 mL tube. Place the 1.5 mL tubes on a 1.5 mL magnetic rack for 1 minute to allow the magnetic particles to separate from the supernatant. Discard the supernatant.
Add 200µL 4°C .
Nucleic Acid Extraction
Warm PM1 from the 55°C for 5-10 minutes before use.
Add 6µL 600µL PM1 to each 1.5 mL tube containing the concentrated sample and Nanotrap Magnetic Virus Particles. Vortex to mix.
Prepare an extraction control by adding 6µL 600µL PM1, and 200µL RNA Shield to an empty tube. Also add 100µL Nanotrap Enhancement Reagent 2 and 150µL Nanotrap Magnetic Virus Particles.
Incubate the samples including the extraction control for 10 minutes at room temperature.
Place the 1.5 mL sample tubes on the magnetic rack and wait 1 minute before transferring the supernatant into the center tube of the rotor adapter.
Place the QIAcube rotor adapters in the QIAcube centrifuge and follow the instructions on the QIAcube control tablet to set up the shaker rack, reagents, and tips. When setting up the reagents, shake to mix the PM5 buffer. Set the elution volume to 100µL. Start the extraction run.
Equipment
| Value | Label |
|---|---|
| QIAcube Connect | NAME |
| Automated nucleic acid extraction | TYPE |
| Qiagen | BRAND |
| 9002864 | SKU |
When the extraction is completed, cap the elution tubes and begin the dPCR steps or store at -80°C if the dPCR run will occur in the following days.
Discard the used pipette tips and wipe the waste drawer and QIAcube workspace with 70% ethanol. After each run, remove the plastic tube holder and the reagent tray before running 2 cycles of UV decontamination.
Detection and Quantification of SARS-CoV-2
Thaw GT-Molecular controls and assay solutions on ice. If necessary, also thaw the extracted RNA on ice. Once thawed, vortex to mix.
Dilute 1µL extracted RNA with 99µL RNase-free water for PMMoV analysis.
Prepare master mixes for PMMoV and N1-N2-BCoV assays. Allow for one extra sample. Vortex to mix. Briefly centrifuge the tubes to collect the master mix at the bottom of the tube.
| A | B |
|---|---|
| Qiagen 4x One-Step Viral RT-PCR Master Mix | 10 |
| Qiagen 100x Multiplex Reverse Transcription Mix | 0.4 |
| GT Molecular N1-N2-BCoV Assay Solution | 2.0 |
| RNase/DNase free water | 7.6 |
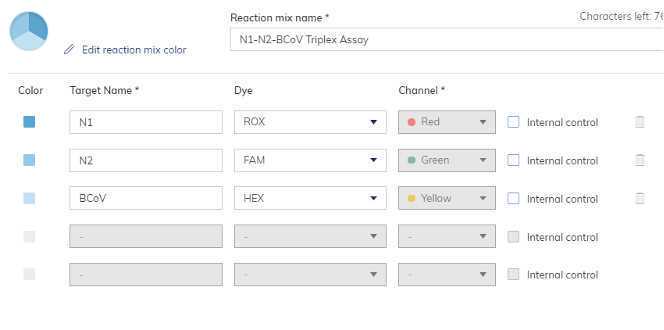
N1-N2-BCoV Master Mix
| A | B |
|---|---|
| Qiagen 4x One-Step Viral RT-PCR Master Mix | 10 |
| Qiagen 100x Multiplex Reverse Transcription Mix | 0.4 |
| GT Molecular PMMoV Assay Solution | 2.0 |
| RNase/DNase free water | 7.6 |
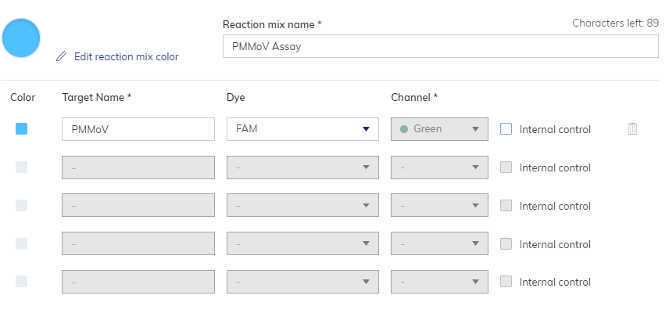
PMMoV Master Mix
Pipette 20µL of the appropriate master mix (N1-N2-BCoV or PMMoV) into the wells of a PCR strip tube.
Add 20µL of extracted RNA sample or positive control to the PCR strip tube following the planned layout. Use the 1:100 diluted samples for the wells being used for the PMMoV assay. After transferring, pipette gently to mix. Keep the PCR strips on ice while loading.
For the non-template control: pipette 20µL of molecular grade water into a PCR tube in place of adding extracted RNA.
Place a Qiagen QIAcuity 26k 24-well Nanoplate onto the Nanoplate protection tray. If the tray is not used, dust can collect on the bottom side of the plate and interfere with the imaging step. Occasionally wipe the tray with 70% ethanol to clean dust.
Using a multichannel pipette, transfer 39µL of solution from the PCR strips to their respective location on the Nanoplate. Be careful to not transfer air bubbles during this step.
Carefully seal the Nanoplate with a Nanoplate seal and the roller provided with the QIAcuity Instrument.
Place the sealed plate in the plate drawer inside the QIAcuity instrument.
Equipment
| Value | Label |
|---|---|
| QIAcuity One, 5-plex | NAME |
| dPCR | TYPE |
| Qiagen | BRAND |
| 911021 | SKU |
Setup the plate by selecting "New Plate". Name the plate and choose the plate type "Nanoplate 26k 24-well".
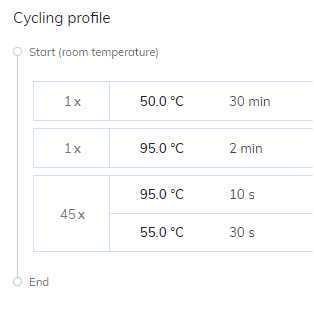
In the dPCR Parameters section under the Priming tab, select the Qiagen Standard Priming Profile.
Navigate to "Samples and controls". Add samples names that are being quantified on this run. Extraction controls should be added as samples. Under the "Controls" tab create both a "N1-N2-BCoV Positive Control" and a "PMMoV Positive Control". Under the Non Template Controls" tab create a "N1-N2-BCoV dPCRNeg" and a "PMMoV dPCRNeg".
Navigate to "Plate Layout". Assign reaction mixes, samples and controls to their wells. Save plate and exit the setup.
On the QIAcuity tablet, select the plate and run the reaction.
Analysis and Interpretations
When the QIAcuity run is complete, ensure the image transfer is marked as complete in the QIAcuity Software Suite before inspecting the data.
Open the plate results by selecting "Analysis". Select all the wells and targets before selecting "Show results".
In the 1D Scatterplot tab verify that the automatic threshold is accurately placed between the negative and positive partitions. If needed, adjust the threshold placement to the accurate position.
Use the "Export to CSV" button in the List tab to export the data.
Index: Preparation of Bovilis Coronavirus
5mL pre-chilled molecular grade water and swirl to mix.
Aliquot 100µL stock in sterile 1.5 mL tubes and label each tube BCoVND (non-diluted). Store BCoVND at-80°C
Dilute 60µL BCoVND with 540µLpre-chilled molecular grade water and pipette to mix. Briefly centrifuge. Label this tube BCoVINT (intermediate).
Dilute 500µL BCoVINT with 49.5mL pre-chilled molecular grade water and invert to mix. Aliquot 1mL of BCoVworking into sterile 1.5 mL tubes and store aliquots at -80°C.
To quantify the BCoV spike, extract the RNA of an aliquot of BCoVworking in triplicate.
Add 100µL of BCoVworking working into three 1.5 mL tubes.
To each tube add 6µL 2-mercaptoethanol and 600µL PM1 from the Qiagen AllPrep PowerViral DNA/RNA Kit and invert to mix.
Microcentrifuge the tubes at 13000x g,0h 0m 0s for 1 minute.
Add the supernatant to the center column of a rotor adapter and continue the extraction in the Qiagen QIAcube.
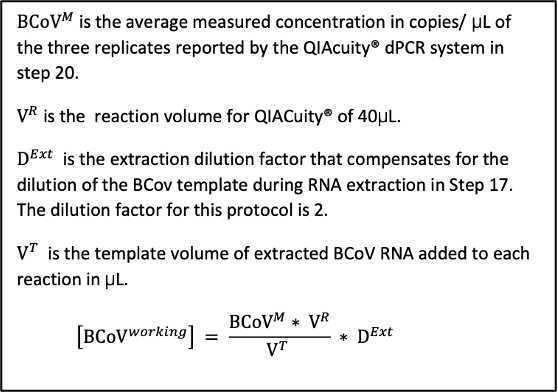
Quantify the extracted BCoVworking RNA by analyzing the extraction triplicates using the same dPCR steps beginning in the Detection and Quantification section .
Index: Imaging Errors
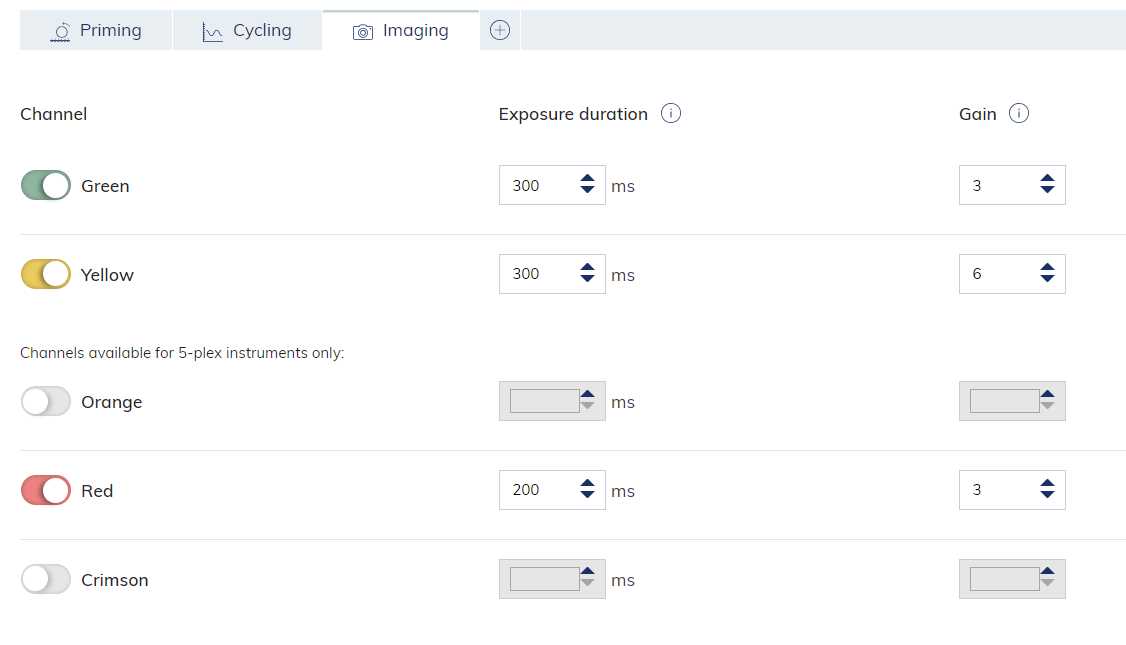
The QIAcuity instrument may show an error when viewing results that a "channel has reached saturation". To resolve this error, reduce the imaging gain and exposure duration and re-image the plate.