Purification of 10xHis-SuperTEV
Kelvin Lau, Bouchra Bouchri, Florence Pojer
Abstract
SuperTEV is a mutated version of TEV (Tobacco Etch Protease) a cysteine protease widely used in labs as it is highly specific to a cleavage sequence that can be genetically encoded. Depending on the lab, it has been reported that the protease is unstable or purifies with low yields. Efforts have been made for many years (Tropea, 2009, Methods Mol Biol) to improve its solubility. Here we report our platform's efforts in generating a new TEV variant, called SuperTEV, that incorporates 9 mutations identified by different groups in recent years applied together at once. We find that the protein is easily produced and purified in large amounts and is functional.
The list of mutations on the canonical TEV protease are the following (with references that also identified the same mutations):
T17S (directed evolution, increased solubility and production) van den Berg et al., 2006 and Wei et al., 2012
L56V (rational design, improved solubility) Cabrita et al., 2007 and Wei et al., 2012
N68D (directed evolution, increased solubility and production) van den Berg et al., 2006 and Wei et al., 2012
I77V (directed evolution, increased solubility and production) van den Berg et al., 2006 and Wei et al., 2012
S135G (rational design, increased solubility) Cabrita et al., 2007 and Wei et al., 2012
I138T (increased catalytic activity; TEV3) Sanchez and Ting, 2019
S153N (increased catalytic activity; TEV3) Sanchez and Ting, 2019
T180A (increased catalytic activity; TEV3) Sanchez and Ting, 2019
S219V (inhibits autoproteolysis) Kapust, 2001
References:
Wei et al., 2012. Protein Expression and Purification
Sanchez and Ting, 2019. Nature Methods
van den Berg et al., 2006. Journal of Biotechnology
Cabrita et al., 2007. Protein Science
Kapust et al., 2002. Biochemical and Biophysical Research Communications
Before start
Recommended to have ready before starting alongside Materials.
-Autoclaved flasks
-Autoclaved LB media
-Autoclaved AutoTB media
-Buffers
-Liquid nitrogen
Steps
Growing bacterial cultures (6 L)
Transform the bacterial plasmid expressing the 10xHis-SuperTEV into BL21 (DE3) cells. Plate on to LB-Agar plates + Kanamycin. Grow overnight at 37°C or over the weekend at 25°C.
In an afternoon, pick a streak of cells and innoculate into 200mL of LB + Kanamycin media. Note: for every 1L of expression culture, you will require 10-20mL of pre-culture. Grow overnight at 37°C
The next morning, innoculate 20-40mL of preculture into 3 flasks containing 2L of Autoinduction TB media (2L in a 5L flask is appropriate). Shake in an incubator at37°C for 3-4 hours until OD600 ~0.8-1. Take a 1 mL sample of the culture. Immediately change the temperature to 18°Cand continue shaking overnight (approximately 20h 0m 0s)
Take a 1 mL sample of the culture and measure the OD600. Harvest the culture by centrifuging at 5000x g,10°C . Transfer the pellets to 50 mL tubes directly or resuspend in minimal amounts wash buffer buffer transferring. Pellets can be used immediately or store at -20°C indefinitely.
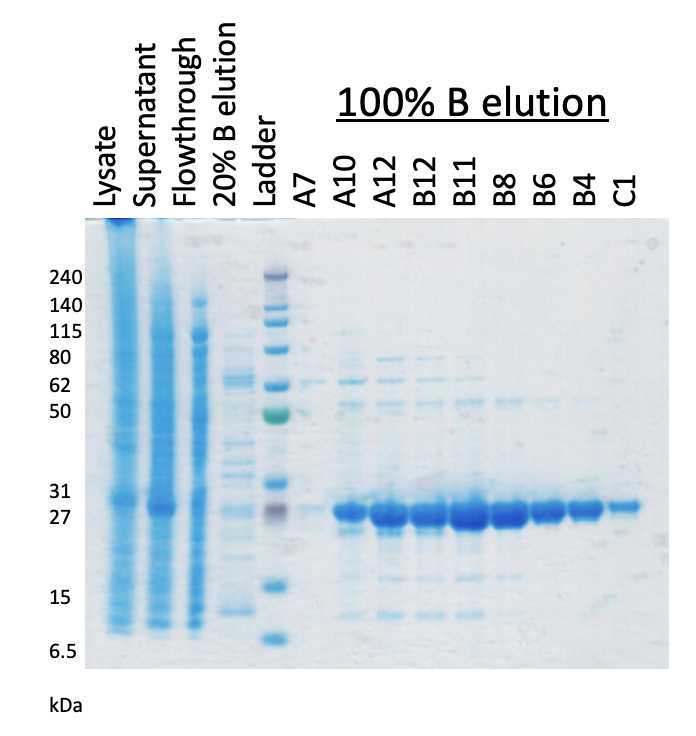
Confirm expression by running an SDS-PAGE gel to observe the appearance of a band around 25 kDa that would represent the production of the 10xHis-SuperTEV.
Recommended recipe to prepare SDS-PAGE samples. Mix 1:4, 4X LDS loading dye and 8 M Urea, to make a Urea loading dye.
For samples, calculate the amount of sample needed to prepare. We use the following formula:
1/OD600 x 100 uL = volume in uL to centrifuge down 14000rpm .
Discard the supernatant, Keep the pellet.
Resuspend pellet in 40 uL of the Urea loading dye.
Load 10 uL on to a NuPage 4-12% bis-Tris SDS-PAGE gel.
Prepare purification buffers
From stock solutions prepare the following :
Filter 0.22 or 0.45 um
| A | B | C |
|---|---|---|
| Buffer | Component | Concentration (mM) |
| Wash buffer (2 L) | NaCl | 700 |
| HEPES, pH 7.5 | 20 | |
| Elution buffer (1 L) | NaCl | 700 |
| HEPES, pH 7.5 | 20 | |
| Imidazole, pH 7.5 | 500 |
Buffer lists for purification
Purification by Ni-NTA on an AKTA system
Resuspended pellets were supplemented with glycerol to 10% and DNase.
The mixture was then lysed using an Emulsiflex device by 3 passes until the lysate was visually not viscous.
Total volume is around100mL. Centrifuge down the lysate at 20000x g,4°C.
Transfer the supernatant to a clean container, being sure not to transfer any liquid that does not contain visually turbid particles near the pellet. Filter the supernatant using a 0.45 um filter
Supplement the filtered supernatant with 1Molarity (M) to a final concentration of 25millimolar (mM)
25millimolar (mM)).
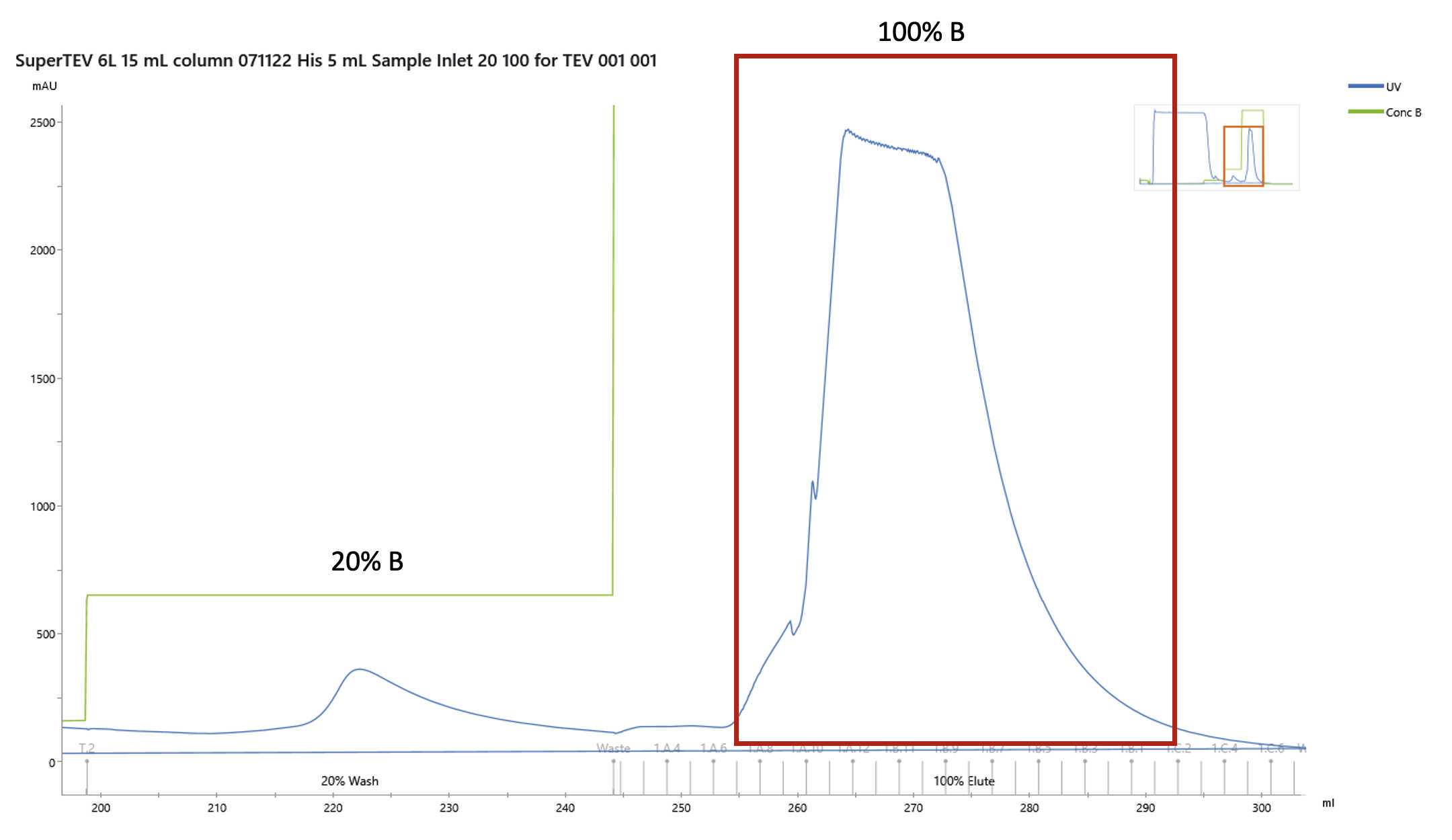
The loaded sample was then washed extensively and eluted as a step gradient as follows
| A | B | C |
|---|---|---|
| Step | Elution buffer concentration (%B) | Column Volumes (CV) |
| Wash + 25 mM imidazole | 5 | 8 |
| Wash + 100 mM imidazole | 20 | 5 |
| Wash + 500 mM imidazole | 100 | 10 |
Gradient for Elution
Collect fractions as appropriate for verification on an SDS-PAGE gel.
The protein elutes at 500millimolar (mM). Fractions containing the purest protein are pooled. It is advised to measure the absorbance at A280 to determine the concentration.
1 mg/mL of SuperTEV = A280, 1.2
Dilute with wash buffer to around 2-3mg/mL or as desired.
Transfer the protein to dialysis bags for dialysis in 2L of dialysis buffer ( 150millimolar (mM), 20millimolar (mM), 5millimolar (mM), 10% (v/v)) at 4C overnight
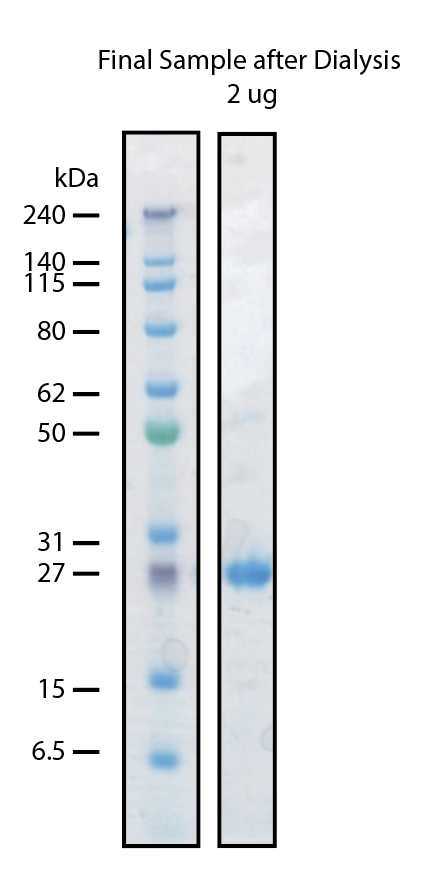
The next day precipitation will be expected. Transfer the dialyzed material to 50 mL tubes and centrifuge 20000x g,4°C. The supernatant should be clear. Once again determine the concentration by measurement at A280.
We have never required to concentrate our TEV protease. We make aliquots that correspond to 1-2mgper tube. Aliquots are flash frozen under liquid nitrogen and then store indefinitely at -80°C
Our final yield is typically in the 10s of mg/L (on average 50 mg/L)
Use of 10xHis-SuperTEV Protease
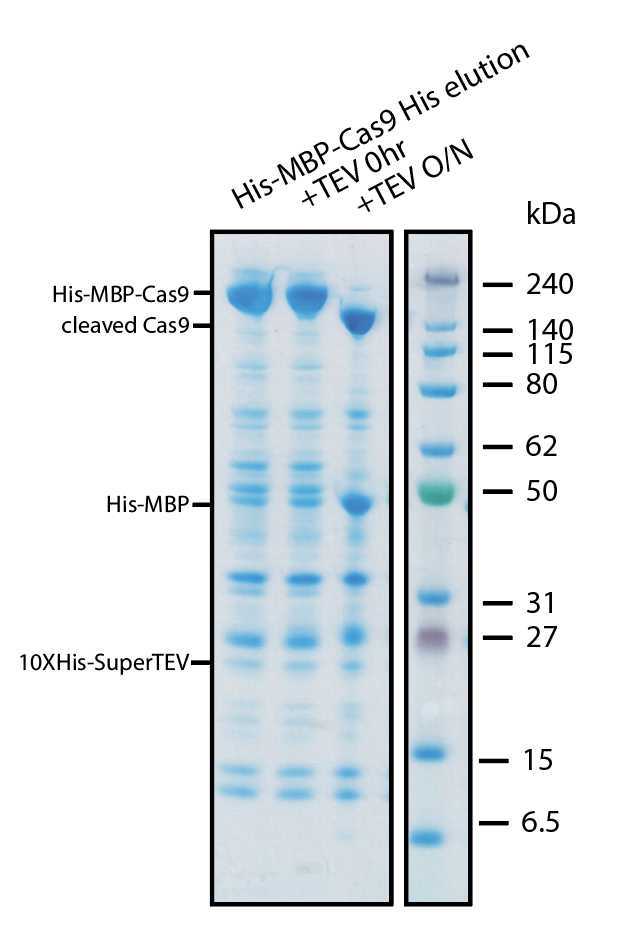
We have tested cleavage of substrates containing the TEV protease site of ENLYFQ/GS. We typically use the protease in a 1:50-1:100 (protease:protein mass ratio). However we have used it also at significantly higher ratios. The protease performs well during dialysis in PBS or HBS buffers atRoom temperature or 4°C