Protocol: Purification of DNA from Whole Blood using the QIAamp Blood Midi Kit (Spin Protocol) + QC with Qubit fluorometer
Kimberley J Billingsley, Breeana Baker, Paula Saffie
Disclaimer
copyright QIAGEN 2013–24.
Thermo Fisher Scientific Inc
Abstract
The QIAamp DNA Blood Midi procedure yield pure DNA ready for direct restriction digestion or amplification. With a suitable centrifuge rotor, eight samples can be prepared simultaneously in about 1.5 hours with approximately 30 minutes of hands-on time.
QIAamp DNA Blood Midi Kits are suitable for use with whole blood that has been treated with EDTA, citrate, or heparin, and samples may be fresh or frozen. Separation of leukocytes is not necessary. The procedure require no phenol/chloroform extraction or alcohol precipitation and involve
minimal handling.
DNA is eluted in Buffer AE or water and is ready for direct addition to PCR or other enzymatic reactions, or it can be safely stored at –30 to –15°C for later use.
The purified DNA is virtually free of protein, nucleases, and other contaminants or inhibitors of
downstream applications.
DNA ranges in size up to 50 kb , with fragments of approximately 30 kb predominating .

Before start
■ Equilibrate samples to r oom temperature (15–25°C) before starting.
■ Prepare a 70°C water bath ( critical initial step and must precede all others in the protocol, Failure to execute prior to proceeding may compromise the integrity and validity of the entire procedure).
■ Ensure that Buffer AW1, Buffer AW2, and QIAGEN Protease have been prepared (Detailed on "materials").
■ If a precipitate has formed in Buffer AL, redissolve by incubating at 56°C.
Attachments
Steps
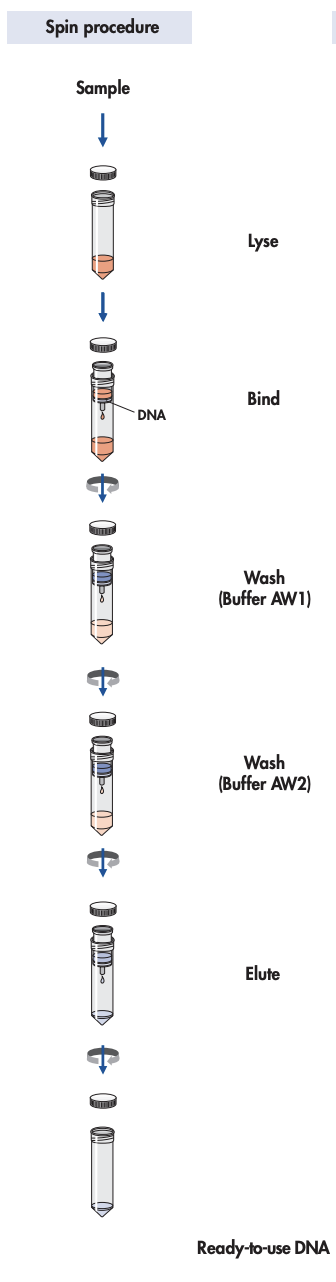
DNA extraction procedure
Pipet200µL QIAGEN Protease into the bottom of a 15 ml centrifuge tube. Add 2mL Sample and mix briefly.
Add Buffer AL 2.4mLBuffer AL, and mix thoroughly by inverting the tube 15 times, followed by additional vigorous shaking for at least 1 min (Check that the tubes are tightly closed).
Incubate at 70°C 10 min.
(During this time, store Protease in the fridge)
Carefully transfer (just squirt solution in middle of the membrane, don't need to go inside the tube with the tip) one half (aprox2-3mL ) of the solution onto the QIAamp Midi column placed in a 15 ml centrifuge tube, cerefully not to moistening the rim .
With the cap tightly closed, centrifuge 3000rpm,0h 0m 0s (1850 x g) for 3min.
Add ethanol 2mLethanol (96–100%) to the sample, and mix by inverting the tube 10 times, followed by additional vigorous shaking.
Remove the QIAamp Midi column, discard the filtrate (dab the rim to remove any exccess of liquid), and place the QIAamp Midi column back into the 15 ml centrifuge tube. Load the remainder of the solution onto the QIAamp Midi column.
With the cap tightly closed, centrifuge 3000rpm,0h 0m 0s (1850 x g) for 3min.
Remove the QIAamp Midi column, discard the filtrate , and place the QIAamp Midi column back into the 15 ml centrifuge tube.
Add Buffer AW1 2mLBuffer AW1 (carefully, without moistening the rim) to the QIAamp Midi column.
With the cap tightly closed, centrifuge at 5000rpm,0h 0m 0s( 4500 x g) for 1 min.
Add 2mL Buffer AW2 (carefully, without moistening the rim) to the QIAamp Midi column.
With the cap tightly closed, centrifuge at 5000rpm,0h 0m 0s(4500 x g) for 15 min.
Place the QIAamp Midi column in a clean 15 ml centrifuge tube , and discard the collection tube containing the filtrate.
Pipet Buffer AE 300µLBuffer AE directly onto the membrane of the QIAamp Midi column and close the cap.
Centrifuge at 5000rpm,0h 0m 0s(4500 x g) for 2 min.
Pipet Buffer AE 300µLBuffer AE directly onto the membrane of the QIAamp Midi column and close the cap.
Incubate at room temperature for 5 min .
Centrifuge at 5000rpm,0h 0m 0s(4500 x g) for 2 min.
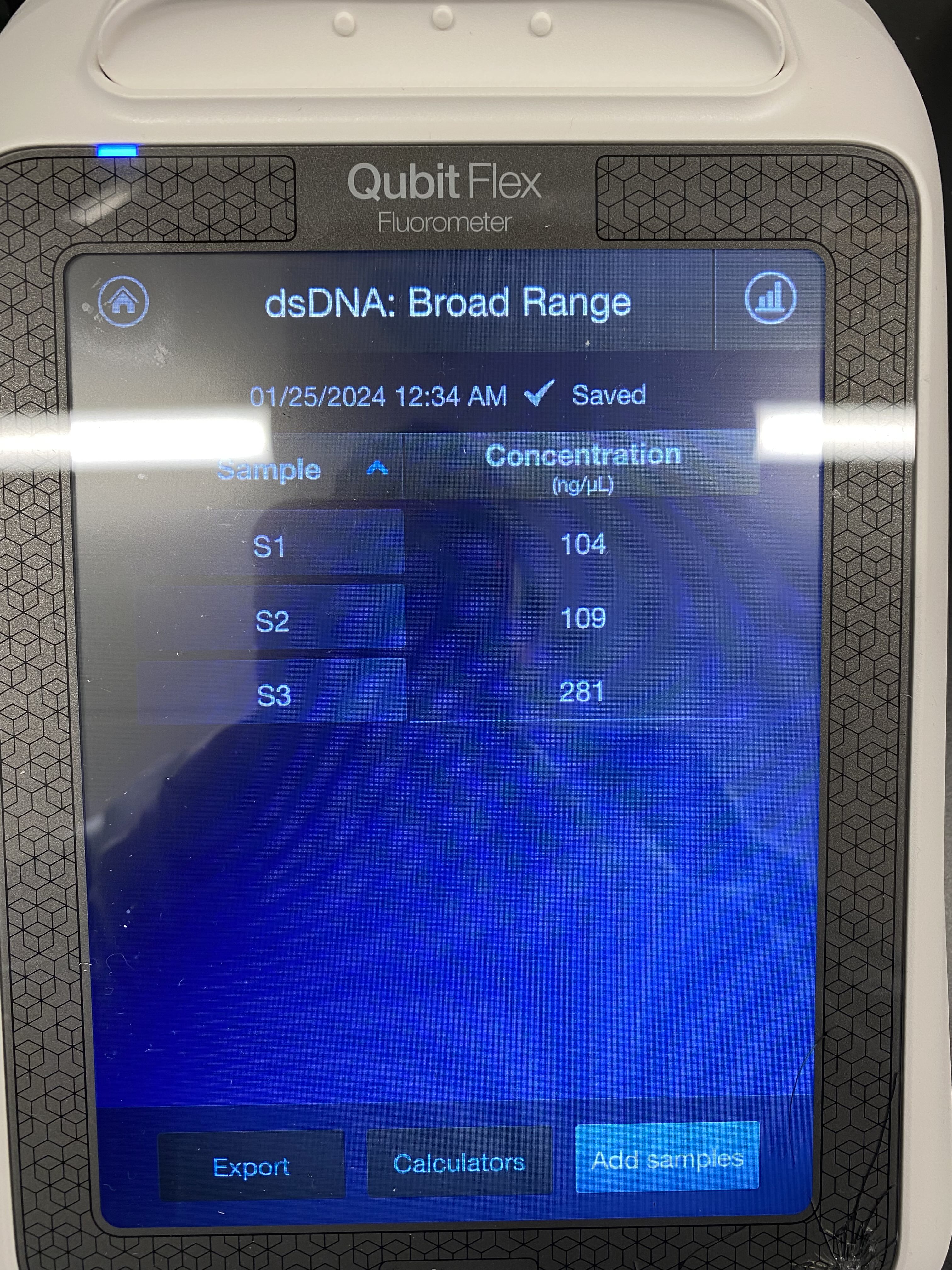
Sample QC procedure (Qubit Fluorometer)
STANDARDIZE THE QUBIT (every time you use it or once a week according to lab protocol)
Get0.2mL Qubit tubes.
If your calibration is successful, press Next to proceed to “Read samples” or begin in this step if it is already calibrated.
In one strip, add10mL of BR Standard 1.
In the other strip, add10mL of BR Standard 2.
Add 190µL of the working solution to all of the tubes.
Vortex, spin down, and incubate in the dark for about 2 min to optimize the reaction.
When prompted, load the QubitTM Flex Tube Strip containing Standard #1 into
the sample chamber, then press Run standards. The reading takes ~3 seconds.
When prompted, load the QubitTM Flex Tube Strip containing Standard #2 into
the sample chamber, then press Run standards. The reading takes ~3 seconds.
READ SAMPLES
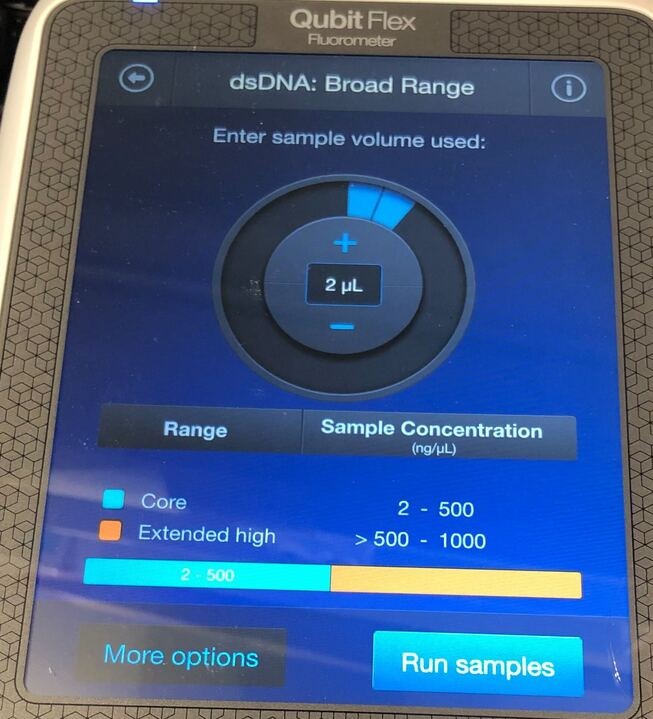
Add198µL or 199µL of dsDNA Broad Range Working Solution to a 0.2mL Qubit tube.
Add2µLor1µLof Samplethoroughly mixed (flick + spin down) in the Qubit tube (200µL total amount )
Pulse-vortex and spin down. Incubate in dark for at least 2min.
Load the tube strip containing the samples as shown. If you have fewer than 8 samples, press to deselect the tube positions that do not contain a sample.
Press Run samples. The reading takes approximately 3 seconds.