Processing frozen human blood samples for population-scale SQK-LSK114 Oxford Nanopore long-read DNA sequencing SOP V2
Kimberley J Billingsley, Laksh Malik, Pilar Alvarez Jerez, Cornelis Blauwendraat, on behalf of the CARD Long-read Team, Abigail Miano-Burkhardt
Long-read sequencing
Oxford Nanopore sequencing
High molecular weight DNA extraction
DNA extraction
Human blood extraction
Whole blood extraction
DNA size selection
DNA shearing
Disclaimer
In development
We are still developing and optimizing this protocol.
Abstract
Abstract:
As part of the GP2 initiative we will generate long-read sequencing data for ~1000 samples to better understand the genetic architecture of Parkinson's disease. To generate this large-scale Nanopore data we have developed a protocol for processing and long-read sequencing frozen human blood samples, targeting an N50 of ~30kb and ~30X coverage.
Acknowledgements:
We would like to thank the Nanopore team (Androo Markham & Jessica Anderson), PacBio team (Jeffrey Burke, Michelle Kim, Duncan Kilburn & Kelvin Liu) and the whole CARD long-read team listed below => UCSC: Benedict Paten, Trevor Pesout, Paolo Carnevali, Mira Mastoras, Melissa Meredith, Jean Monlong, Ryan Lorig-Roach, Mobin Asri; NE: Miten Jain; NCI: Mikhail Kolmogorov; NHGRI: Adam Phillippy, Arang Rhie; Baylor: Fritz Sedlazeck, Farhang Jaryani; JHU: Winston Timp; NIA: Cornelis Blauwendraat, Kimberley Billingsley, Pilar Alvarez Jerez, Laksh Malik, Breeana Baker, Maysa Abdelhalim, Kensuke Daida, Rylee Genner, Abigail Miano-Burkhardt, Caroline Pantazis; This protocol was optimized using frozen blood samples from the PPMI initiative. PPMI – a public-private partnership – is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, a full list of the PPMI funding partners can be found at www.ppmi-info.org/fundingpartners.

Steps
Part 1: Preparing Blood Samples (~30 min for 24 samples)
Obtain blood samples from -80C freezer and thaw in water bath at 37°C for 0h 15m 0s
Note: This is specifically for 6mL tubes, if starting with only 1mL thaw until warm (~ 0h 5m 0s)
Inversion mix blood 10x immediately before use.
Note: The blood sample needs to be very thoroughly mixed in order to ensure better DNA isolation.
Note: We will be taking 1mL forward. If your samples have a volume larger than 1mL, aliquot 1mL out into 1.5mL Eppendorf DNA LoBind tubes and keep frozen at -80°C until ready to use. It is important to limit the amount of freeze/thaw cycles as much as possible, the protocol will work best at the first freeze/thaw cycle.
Part 2: KingFisher Apex Nanobind HMW DNA Extraction protocol (~3 hours for 24 samples)
Note: Protocol is adapted from PacBio Nanobind HT HMW DNA Extraction 1mL Whole Blood KingFisher Apex protocol: https://www.pacb.com/wp-content/uploads/Procedure-checklist-Extracting-HMW-DNA-using-Nanobind-HT-1-mL-blood-kit-for-mammalian-whole-blood-on-KingFisher-Apex-system.pdf
Note: The KingFisher Apex protocol can be run with up to 24 samples at once.
We will be using the Nanobind HT 1mL Blood Kit (102-762-800) from PacBio.
Prepare the KingFisher 24 deep well plates as follows:
Plate 1: Elution: 200µL Buffer EB
Plate 2: CW2 Wash 1: 2000µL Buffer CW2
Plate 3: CW2 Wash 2: 2000µL Buffer CW2
Plate 4: CW1 Wash: 2000µL Buffer CW1
Plate 5: Nanobind storage: 5mm Nanobind Disks
Plate 6: Lysis Binding: 100µL Proteinase K + 1000µL Whole Blood + 100µL RNase A + 750µL Buffer BL3
Note: Add BL3 gently against side of well. Adding BL3 directly to the solution may affect extraction performance.
Plate 7: Comb tip: 24 Flex tip comb
Note: Sample and reagents in lysis binding plate must be added to the plate in the order listed above.
Run Kingfisher program: 1mL_Blood_Nanobind_HT_APEX script (102-998-100).
After ~0h 50m 0s when the program pauses, add 1500µL isopropyl alcohol (IPA) to lysis binding plate.
Note: Add IPA gently against side of well. Adding IPA directly to solution may affect extraction purity.
When the program ends after ~1h 45m 0s (from the start), transfer eluate to a 1.5mL Eppendorf DNA LoBind tube.
Note: The program is designed to leave the nanobind disk in the elution plate. If the nanobind disk ends up in the tip comb plate, this does not affect extraction performance. Use a P200 to remove any elution buffer remaining on the nanobind disk.
Pipette mix samples 10x to homogenize.
Let samples rest overnight at room temperature (RT).
After overnight rest at RT, DNA can be stored at 4°C for up to four weeks, or -80°C indefinitely.
Part 3: Pre-Size Selection DNA Quantification (~30 min for 16 samples)
Note: This duration does not include the time it takes to size the samples. The total time it takes to size 16 samples on the TapeStation or the Femto Pulse is ~45 minutes or ~3 hours and 30 minutes, respectively.
Following overnight rest, pipette mix samples 10x.
Quantify by taking a single measurement on the Qubit Flex Fluorometer.
Size on the Agilent Tapestation 4200 or Agilent Femto Pulse System. The expected size range for samples post-extraction is >80kb.
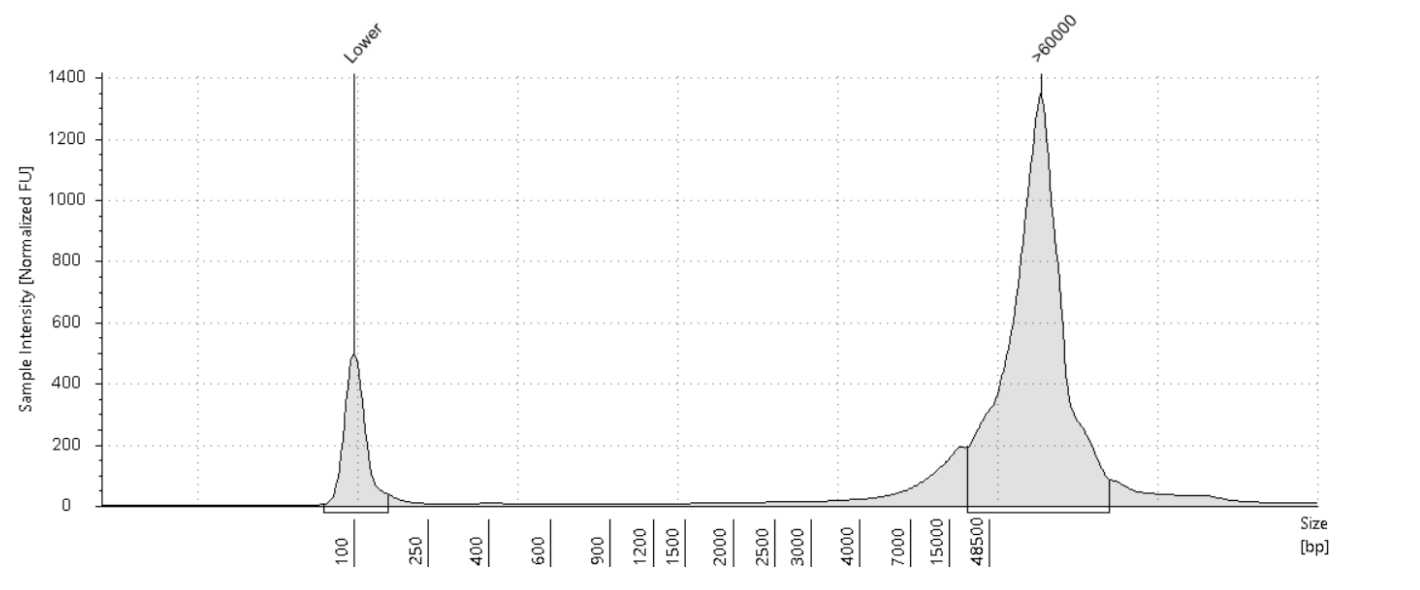
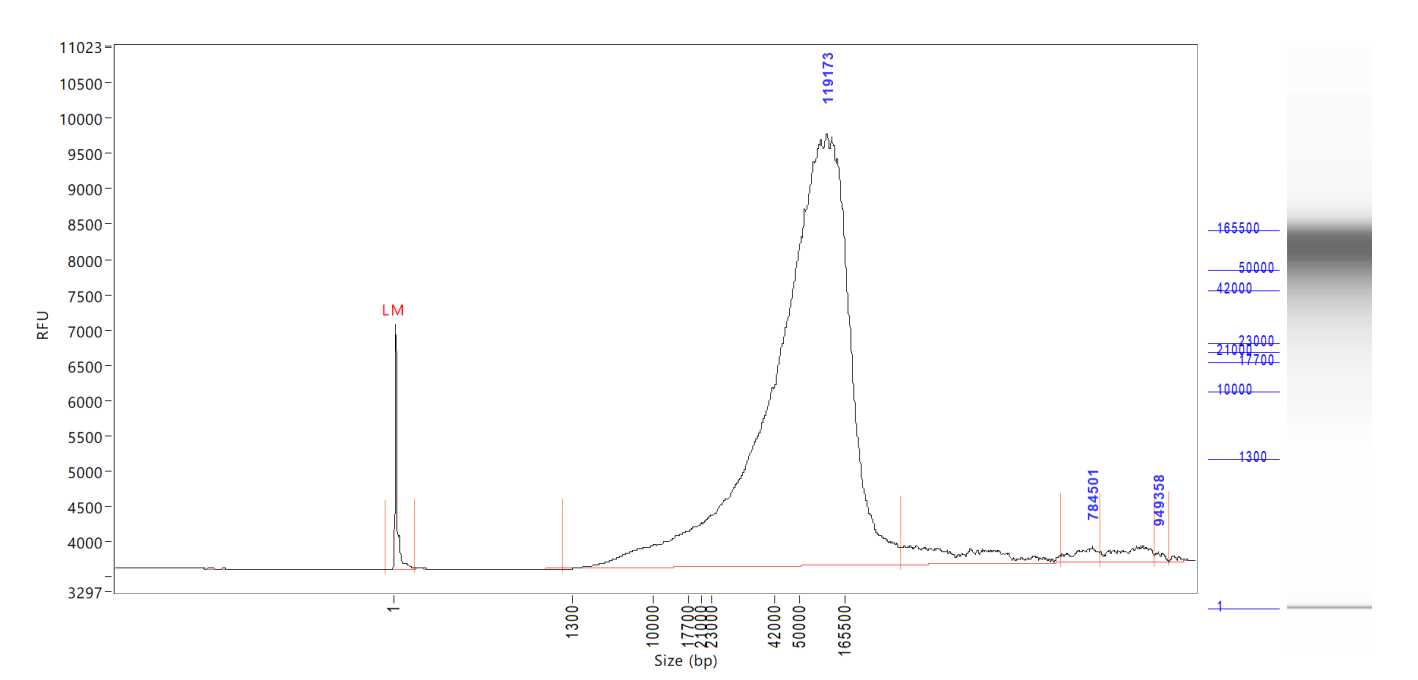
Note: I t is recommended to use the Femto Pulse System to get a better representation of the size distribution of the DNA samples.
Once all the samples are properly quantified, move on to size selection.
Note: Blood DNA recoveries after KingFisher are very variable per sample and per run. If recovery is very low (<5ug) repeat Parts 1-3 with 2 x 1mL of sample in question. If either of the extra runs has a good recovery, move on with that DNA. If all three are bad, you can pool them together to reach DNA target amount.
Part 4: Size Selection (~2 hours for 16 samples)
Note: This step was originally optimized for R.9 which required more input DNA. R.10 requires a lower DNA input amount so for best results we recommend first shearing your samples and then size selecting, either using a Blue Pippin or manually with the PacBio Short Read Eliminator Kit (102-208-300).
Note: If you have to pool multiple 1mL runs for a sample, it is possible that the concentration will be <40ng/uL. If it is lower, a clean up step here may be used to re-elute the pool into a smaller volume to increase the concentration.
Ensure your samples are between 40-150ng/uL. More concentrated samples will need dilution with TE buffer or water. Size selection is not recommended for samples below 40ng/uL.
Note: TE buffer is preferred for long-term storage since it reduces DNA degradation over time.
While Set 2 is spinning, carefully remove the supernatant without disturbing the DNA pellet from Set 1. Pellet may not always be visible but will have formed on the bottom of the tube under the hinge region. Pipette from the opposite wall (thumb lip side of tube).
For very viscous DNA, the pellet may not initially form tightly and can lead to easy aspiration of DNA. If this is the case, remove as much supernatant as possible but leave behind what you cannot pipette without aspirating DNA.
Note: Pellet will be tighter after the ethanol (EtOH) wash.
Carefully add 200µL of fresh 70% EtOH on the opposite side of the hinge wall. Remove Set 2 from the centrifuge and place Set 1 in the centrifuge. Centrifuge at 10,000 x g for ~0h 5m 0s at 25°C.
Note: Do not mix after adding EtOH.
While Set 1 is spinning, carefully remove the supernatant without disturbing the DNA pellet from Set 2.
Carefully add 200µL of fresh 70% EtOH on the opposite side of the hinge wall. Remove Set 1 from the centrifuge and place Set 2 in the centrifuge. Centrifuge at 10,000 x g for ~0h 5m 0s at 25°C.
Carefully remove EtOH wash from Set 1 without disturbing the pellet.
Add 50-100uL of Buffer EB and let incubate at RT for 0h 20m 0s.
Note: Letting the sample incubate for longer may better homogenize the sample and may give better quantification readings.
Repeat steps 4.15 and 4.16 for Set 2 once it is done spinning.
After incubation, flick the tube to ensure proper mixing.
Start a microcentrifuge at 10,000 x g and 25°C for 0h 5m 0s while preparing the samples.
In the meantime, place the necessary sample volume for at least 6ug into a 1.5mL Eppendorf DNA LoBind tube.
Note: Expect to lose 40-60% of material during size-selection. Do not take anymore than 20ug into size selection.
We will be using the PacBio Short Read Eliminator Kit (102-208-300).
To each sample, add equal volume of Buffer SRE.
Mix thoroughly by flicking the tube.
Load tube into the microcentrifuge with hinge facing out and make sure samples are balanced.
Note: Inserting tube with the hinge out is crucial to avoid aspirating the pellet if not visible in later steps.
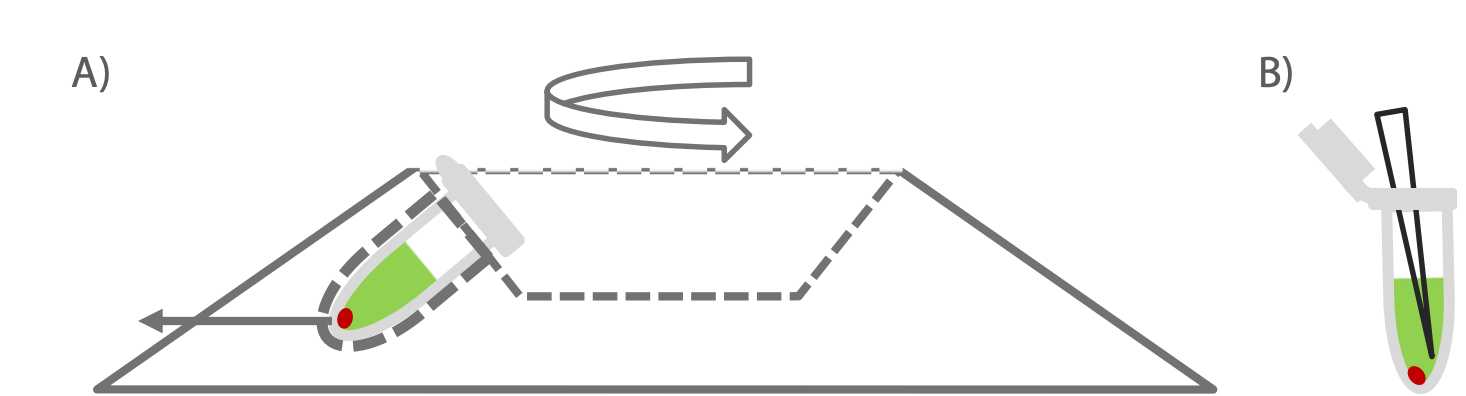
Centrifuge at 10,000 x g for 0h 30m 0s at 25°C.
During this time, prepare 250µL per sample of fresh 70% EtOH in nuclease-free water.
Remove the first set of eight tubes (Set 1) from the centrifuge, repositioning the second set of eight tubes (Set 2) in the centrifuge so they are balanced. Spin Set 2 at 10,000 x g for another ~0h 5m 0s at 25°C.
Part 5: Post-Size Selection DNA quantification (~10 min for 8 samples)
Analyze size selection recoveries via Qubit.
Note: Quantification post-size selection can be difficult. If necessary, gently pipette mix the sample 10x before quantification to homogenize. The most accurate quantification will come after Megaruptor shearing in Part 6, but an estimate of DNA concentration is important in this step to standardize the samples going into shearing.
Part 6: Shearing (~3 hours for 8 samples)
We will be using the Diagenode Megaruptor 3 Shearing Kit (E07010003).
In a Megaruptor 3 shearing tube, make up the size selected sample to 150µL and 40-70ng/uL with TE Buffer or water.
Note: It is ok if target concentration cannot be reached for all samples. If concentration differs from this range, make sure the Megaruptor 3 shearing settings are updated to reflect your sample concentration. If shearing more than one sample try to get all the concentrations to be as close as possible to this target range and/or to each other.
Note: We recommend taking in a larger concentration into shearing since you will lose some sample when removing the syringe post-shear. If the sample concentration is higher than 70ng/uL post-SRE but diluting it with TE buffer or water to 150uL would make it less than 70ng/uL, it is suggested to dilute to 100uL rather than 150uL. If the sample concentration does not reach 70ng/uL post-SRE, it is suggested to take all sample into shearing and do not dilute with TE buffer or water. Make sure Megaruptor 3 shearing settings reflect your exact sample volume.
Attach the Megaruptor 3 shearing syringe onto the tube and push the entire item into the MR3 slots until it fits snugly. If running fewer than 8 samples, put the tubes in the 1st and/or 8th slots, working your way in.
Note: Samples should always be balanced. If running an odd number of samples, samples can be balanced with an empty corresponding tube.
Shear twice at speed 20 (takes ~3h 0m 0s if the MR3 is set to 150uL volume).
Once finished, remove the sample from the Megaruptor 3 and carefully remove the syringe from the tube. Make sure the plunger is fully depressed in order to avoid losing sample. Use a P200 pipette to aspirate any leftover sample on the syringe.
Avoid any vortexing of the DNA from this point on to avoid any unnecessary further shearing. Instead mix by gently flicking the tube and spin down.
Part 7: Post-Shear DNA Quantification (~10 min for 8 samples)
Quantify the samples on the Qubit and size using the Agilent TapeStation 4200 or the Agilent Femto Pulse System. Although the targeted size is 30kb, the observed size range for samples post-shear is between 30-70kb. These usually still sequence around 30kb.
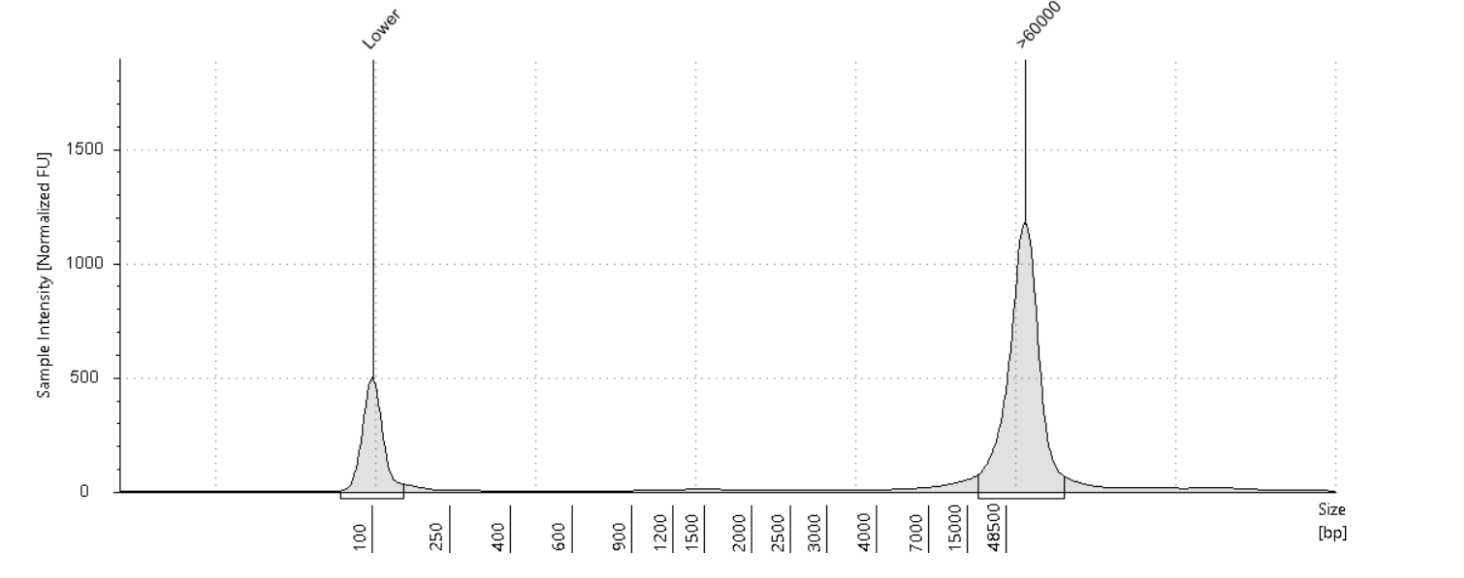
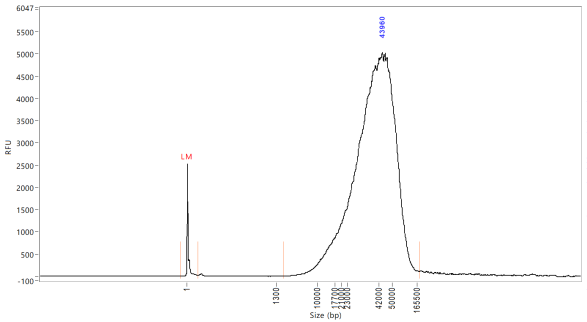
Upload the TapeStation/Femto reports and quantifications to tracking google sheet.
At this point, at least 2.5µg of DNA is necessary to move on to library prep.
The DNA can be stored at 4°C for up to four weeks, or -80°C indefinitely.
Part 8: Manual SQK-LSK114 Library Prep and Sequencing (~6 hours, including reloads)
Note: Library prep can also be done on the Hamilton NGS Star and can process 48 samples in ~4 hours using the HAMILTON NGST STAR Oxford Nanopore SQK-LSK114 Library preparation SOP: https://dx.doi.org/10.17504/protocols.io.n2bvj36mnlk5/v1
Note: Reagents are from Oxford Nanopore Ligation Sequencing Kit V14 (SQK-LSK114). Library prep is following the standard Oxford Nanopore Ligation Sequencing DNA V14 (SQK-LSK114) protocol with minor tweaks: https://community.nanoporetech.com/docs/prepare/library_prep_protocols/genomic-dna-by-ligation-sqk-lsk114/v/gde_9161_v114_revo_29jun2022?devices=promethion
Minor tweaks:
- 2.5ug DNA library in
48µL(Part A Step 2) - Add
45µLof AMPure XP beads (Part B Step 10) - Use Short Fragment Buffer (Part B Step 14)
- Incubate for
0h 20m 0sat37°Cand 300-450 x g (Part B Step 18)
A. DNA Repair and End-Prep
- Place all the necessary reagents on ice to thaw and the Agencourt AMPure XP beads out at room temperature.
- Prepare the following in a 0.2mL thin-walled PCR tube:
48µLDNA (input 2.5ug, this might be over 48uL but that is fine. Adjust the amount of beads to match the total volume of this mixture (sample + buffers/enzymes))3.5µLNEBNext FFPE DNA Repair Buffer (vortex and spin down)3.5µLUltra II End-Prep Reaction Buffer (vortex and spin down)3µLUltra II End-Prep Enzyme Mix (do not vortex, spin down)2µLNEBNext FFPE DNA Repair Mix (do not vortex, spin down)
Note: In our experience, AMPure XP beads and Short Fragment Buffer perform best when thawed and used at RT rather than on ice.
Note: Do not exceed 180uL for total volume.
-
Mix thoroughly by gently flicking the tube or very gently pipetting up and down 10x, and then spin down.
-
Using a Thermocycler, incubate samples at
20°Cfor0h 30m 0sand then65°C
for 0h 5m 0s.
Note: Start and pause Thermocycler to allow lid to come to 85°C before putting samples in.
-
Resuspend the AMPure XP beads by vortexing.
-
Transfer DNA samples to clean 1.5mL Eppendorf DNA LoBind tube.
-
Add
60µL(or equivalent volume, see step 2) of resuspended beads to the reaction and mix by flicking the tube 10x. Do not pipette mix here as beads may clump around the pipette tip. -
Incubate on a rocking laboratory shaker or hula mixer for
0h 5m 0sat RT. -
Prepare
500µLper sample of fresh 80% ethanol in nuclease-free water. -
Spin down and pellet sample on magnet until eluate is clear and colorless, about
0h 2m 0s -
Keep the tube on the magnet and pipette off the supernatant.
Note: Can retain if needed just in case the final elution quantification is uncharacteristically low.
- With the samples remaining on the magnet, wash the beads with
200µLof 80% EtOH, pipetting on the opposite wall making sure not to disturb the pellet. Once EtOH has been added to all samples, immediately remove and discard the EtOH.
Note: The goal here is to make sure the beads are fully covered. If initial volume of beads was significantly higher than 60uL, more EtOH may be required.
-
Repeat step 12.
-
Spin down and place the tube back on the magnet, pipetting off any residual EtOH.
-
Allow to dry for ~
0h 0m 30sbut do not overdry to the point of cracking. -
Remove the tube from the magnetic rack and resuspend the pellet in
62µLnuclease-free water. Incubate for0h 3m 0sat RT, gently flicking every so often. If not quantifying the sample post-DNA repair and end-prep, user can resuspend the pellet in 60uL water. -
Spin down and pellet the samples on a magnet until eluate is clear and colorless.
-
Remove and retain
62µLof eluate into a 1.5mL Eppendorf DNA LoBind tube. -
Optional: Quantify 2uL of sample on the Qubit.
Note: It is possible to store samples at 4°C overnight at this step if necessary.
B. Adapter Ligation and Clean-Up
- Spin down the Ligation Adapter (LA) and NEBNext Quick T4 DNA Ligase, then return to ice.
Note: Do not allow LA or Quick T4 to remain at room temperature for too long. Since LA and Quick T4 do not freeze at -20°C, it is possible to leave them in the freezer until needed.
-
Thaw Ligation Buffer (LNB) at RT, mix by pipetting up and down (vortexing is ineffective due to viscosity), and place on ice.
-
Thaw Elution Buffer (EB) and Short Fragment Buffer (SFB) at RT, mix by vortexing, spin down, and place on ice.
-
In a 1.5mL Eppendorf DNA LoBind tube, mix the following in order:
60µLDNA sample25µLLNB10µLQuick T45µLLA
-
Mix by gently pipetting and spin down.
-
Incubate the reaction for
0h 30m 0sat RT. -
During this time, put flow cells out at RT.
-
Resuspend AMPure beads by vortexing.
-
Add
45µLof resuspended beads to the reaction and mix by flicking. -
Incubate on a rocking laboratory shaker or hula mixer for
0h 5m 0sat RT. -
Spin down sample and pellet on magnet.
-
Keeping tube on magnet, pipette off the supernatant.
Note: Can retain supernatant if needed just in case the final elution quantification is uncharacteristically low.
-
Wash the beads with
250µLSFB, remove from magnet and flick to resuspend, spin down and re-pellet on magnet, and then remove and discard supernatant. -
Repeat step 14.
-
Spin down and place the tube back on magnet, pipetting off any residual supernatant.
-
Allow to dry for ~
0h 0m 30s, but do not overdry to the point of cracking. -
Remove the tube from magnet and resuspend pellet in
26µLEB, spin down, and incubate for0h 20m 0sat37°Cand 300-450 x g. -
During this time, QC the flow cells.
Note: Wait at least 0h 20m 0s after taking out the flow cells to allow them to reach RT before loading onto the PromethION to avoid condensation formation.
Note: If trying to reach 30x coverage, it is recommended to only use flow cells with >7000 pores.
-
Pellet the beads on a magnet until eluate is clear and colorless.
-
Remove and retain
26µLof eluate into a clean 1.5mL Eppendorf DNA LoBind tube, being careful not to aspirate any beads (this is the DNA library). -
Quantify 2uL of sample on the Qubit.
-
Re-prep library from Part 6 if <50 fmol. We have calculated this based on a DNA size of 30kb and at a coverage of 30x over three loads. To convert ng to fmols, we used https://nebiocalculator.neb.com/#!/dsdnaamt.
Note: 60 fmol will be enough for three loads of 20 fmol each, which is the amount that our testing has indicated is necessary to hit 30x coverage. If necessary, user can load as low as 10 fmol for the third load.
- Keep libraries on ice until ready to load on flow cell.
C. Priming and Loading R10 Flow Cell
Note: This kit is only compatible with R10.4.1 flow cells (FLO-PRO114M).
- Thaw Sequencing Buffer (SB), Library Solution (LIS) or Library Beads (LIB), Flow Cell Tether (FCT), and Flow Cell Flush (FCF) at RT, vortex, and spin down.
- Add
30µLof thawed and mixed FCT directly to tube of thawed and mixed FCF and vortex. Alternatively, in a new tube, add30µLof thawed and mixed FCT to1170µLof thawed and mixed FCF and vortex. This is your priming mix - Rotate valve to reveal inlet port 1 on the flow cell, set P1000 pipette to
200µLand use the dial to draw back a small amount of volume to remove any air bubbles (usually about 20-30 μL, just until a small volume of buffer enters the pipette tip). - Flush
500µLof priming mix into inlet port 1 of the flow cell, being extremely careful to avoid the introduction of air bubbles at the end. - Wait
0h 5m 0s - During this time, make up your DNA library to
32µLat 20 fmol using EB. - Prepare the library mix for loading:
100µLSB68µLLIS or LIB32µLDNA library (20 fmol)
-
Repeat steps 4 and 5.
-
Gently pipette mix the prepared library mix right before loading.
-
Load
200µLof the library mix into inlet port 1 on the flow cell. -
Close valve to seal inlet port and close PromethION door.
-
Wait
0h 10m 0sand then initiate sequencing. -
Ideally, the library quants yielded at least 60 fmol to allow for 3 x 20 fmol loads, the latter 2 loaded approximately after 24 and 48 hours. However this will vary slightly depending on pore usage, data generated, as well as other factors (i.e., if after 24 hours there are still 3000+ pores then the sample does not need to be reloaded until 48 hours).
-
To wash and reload a flow cell, begin by thawing Wash Mix (WMX) on ice and Wash Diluent (DIL) at RT Note: DIL should be vortexed. WMX should NOT be vortexed, only spun.
-
In a new tube, add
2µLWMX to398µLDIL and pipette mix. This is your flow cell wash mix. -
Pause the PromethION runs and export .pdf run reports.
-
With inlet port 1 closed, remove waste from port 2 or 3.
-
Rotate the inlet port 1 cover to reveal inlet port 1.
-
Using a P1000, insert tip into inlet port 1 and draw back a small volume using the wheel to remove any air bubbles (usually around 20-30 μL, just until a small volume enters the pipette tip).
-
Load
400µLflow cell wash mix into inlet port 1, avoiding any introduction of air. -
Wait
1h 0m 0s -
Repeat priming steps and reload samples (steps 1 - 13).
D. Flushing and Recycling Flow Cells (~15 min per set of 4 flow cells)
- Following the completion of the sequencing, flow cells may be removed from the sequencer.
- Place enough absorbent material to take up approximately 4 mL of flush waste.
- Rotate valve to reveal inlet port 1.
- Place flow cell at a 45° angle on the absorbent material and, using a P1000, flush 1 mL of DI water into the inlet port.
- Repeat 3 more times for a total of 4 mL.
- Once complete, close the inlet port cover and remove all liquid from the waste port.
- Dispose of absorbent material as local biological waste guidelines dictate.
- Return flow cells to clear plastic tray in which it was shipped.
- Put the clear plastic lid back on the tray.
- Place the tray back in the packaging.
- Place packaged cells in the returns box (large box can hold up to 80 flow cells or 20 packs).
- Once returns box is filled, follow the instructions here and follow the prompts to request the box to be sent back to Nanopore.



