Preparation of cells for live-cell imaging of a FOXO-based AKT kinase translocation reporter by widefield microscopy
Ralitsa R Madsen
Abstract
This protocol provides detailed instructions for imaging of cells with stable expression of a kinase translocation reporter, alongside a nuclear marker for segmentation. We have used this approach to measure the nuclear-cytoplasmic shuttling of a FOXO-based AKT reporter in both HeLa cells and human induced pluripotent stem cells (iPSCs). Note that we culture our cells without antibiotics except during live-cell imaging as indicated.
For subsequent data analysis, please refer to the pipeline provided on the Open Science Framework under doi: 10.17605/OSF.IO/4F69N.
Before start
Prepare coating solution: thaw Matrigel stock aliquot on ice, then dissolve 400 µl in 19.6 ml of cold OptiMEM * Prepare autoclaved forceps
- Prepare complete medium (CM) in advance, e.g. for HeLa cells (see materials for individual items): DMEM with L-Glutamine and Sodium Pyruvate (500 ml) + L-Glutamine (5 ml) + FBS (50 ml) +/- selection antibiotic for stable cell lines
- Prepare live cell imaging (LCI) medium: 49 ml Fluorobrite DMEM + 500 µl L-Glutamine + 500 µl Penicillin-Streptomycin
- Make sure that the microscope, including objectives, holders and oil are warmed up to 37 degrees Celsius for least 3 hours prior to set up for imaging
- Assemble the fluidics system by threading the end of a 40-50-cm-long piece of PFTE tubing into a 1.5-2 cm-long piece of Pharmed tubing with a luer lock at the other end. Roll the tubing and store away in a 150 cm putri dish when not in use. When in use, the end of the PFTE tubing that is free must be threaded through an appropriately sized hole in the lid of an imaging dish - this hole can be made using a flame to heat a needle, then melting the plastic in the lid until the required size has been reached.
Steps
Dish assembly and coating
Take the required number of Ibidi dishes that will be needed for the experiment, along with the equivalent number of inserts. (NB: you can reuse inserts once provided that they have been sterilised in 70% ethanol and dried since the last use; we do this by passing them through PBS once, then 2x across two distinct 70% ethanol aliquots in 50 ml Falcons, then transfer to dry in a closed Petri dish).
Using autoclaved forceps, pick up the individual inserts and place an insert in each dish. Invert the dish once to check that the insert is sticking to the bottom of the dish.
Label the lid of the dish with the well layout. To ensure that you can keep track of the orientation even when you remove the lid, label both lid and base with "N" (north) and "S" (south) in the equivalent positions (see attached schematic).
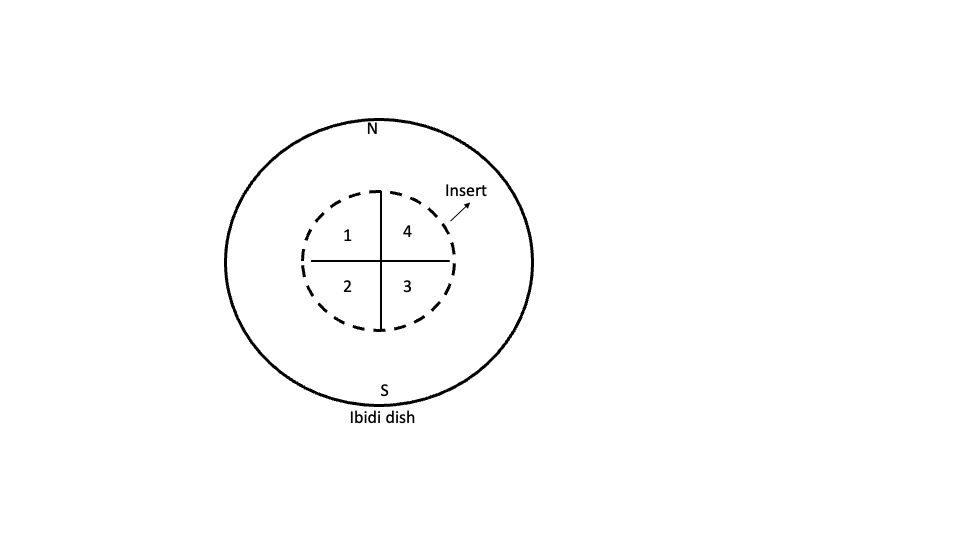
Using ice-col Matrigel solution (1:50 dilution in OptiMEM, prepared in advance), coat each well with 100µL.
5mL Note the coating time and transfer the dishes to the incubator for a minimum of 0h 45m 0s (NB: we usually seed within 1-2h of coating).
Make sure to keep the Matrigel solution on ice and return to the fridge when done.
Cell seeding (the given volumes for splitting are for HeLa cells in T25 flasks)
Take the cells for processing out of the incubator and confirm healthy state by light microscopy examination. The cells should be 80-90% confluent.
Remove the spent medium and wash the cells with 5mL DPBS.
Remove the DPBS and add 0.75mL TrypLE Express.
Incubate the cells for 0h 8m 0s at 37 degrees Celsius.
If you are processing more than one cell line (e.g. different HeLa mutants), proceed with the next, ensuring that you stagger individual cultures 2 minutes apart. DO NOT process multiple cell lines for splitting the same time as this poses a cross-contamination risk.
Once the cells have dissociated, add3.3mL of complete medium (CM), resuspend the cells and transfer to an appropriate labelled 5 ml Eppendorf tube. The final volume should be approximately 4 ml.
Immediately after transfer, mix 20µL of the cell suspension with 20µLTrypan Blue.
Count the cells using a haemocytometer (NB: we find this method to be the most reliable compared to automated cell counting)
Dilute the cell suspension to 30000 cells/ml (NB: ensure sufficient mixing at all stages, including of the stock cell suspension) in a final volume of 1mL (use 1.5 ml Eppendorf tubes).
Take out the coated dishes from the incubator.
For each well to be seeded, take the respective cell suspension and mix 2-3 times with a P1000 Gilson pipette set at 500 µl.
Immediately after mixing the cell suspension, aspirate the coating solution from the well to receive cells using non-filter tips (be careful not to let the wells dry out, yet also take care to observe sterile techniques!).
Transfer 100µL (3000 cells) of the mixed cell suspension to each well. Make sure not to tilt the dish at any point after cell seeding. (NB: if you left the mixed cell suspension sit for more than 1-2 minutes, mix again before seeding though be mindful of time as the wells dry out once the coating has been removed).
NB: We have tested a range of densities and confirmed that the results are reproducible across low- and high-density cultures. The range we have tested is 2500 cells/well up to 7500 cells/well.
Repeat this procedure for the remaining cell suspensions.
Once you have seeded all wells, leave the dishes in the tissue culture cabinet for 15-20 minutes before transfer to the cell culture incubator. This ensures a more even seeding distribution.
Leave the cells in the cell culture incubator for 20-28 hours after seeding.
Serum-starvation
Approximately 24 hours following seeding, inspect the cells under the light microscope to check health, then transfer to the tissue culture cabinet.
Using autoclaved forceps, remove the insert from each dish (NB: either discard if it has been used once before, or sterilise as described in Step 1).
Using non-filter tips, aspirate the spent medium from the cells from the area where the insert boundary is visible (do not touch the cells).
Gently add 3mL of live cell imaging (LCI) medium to the dish, taking care to cover the inner circle containing the cells. This is a wash step to ensure phenol red and serum removal.
Remove the LCI wash and replenish the cells with exactly 2mL LCI, then return to the incubator for 2h 30m 0s prior to microscopy set-up.
Unlike for TIRF, for the wide field imaging of the kinase-translocation reporters, we can image up to 4 dishes at once. It is therefore possible to serum-starve all four dishes at once, however, we recommend that this is done sequentially to avoid any cell areas from drying out while aspirating and replenishing media solutions.
Remember to switch on the microscope temperature unit on at 37 $$°C at least 2h in advance of initiating imaging, and ensure that all components (including lens and dish holders) are heated up.
Preparation of treatment solutions
During the 2.5h wait, prepare the treatment solutions in LCI following the example tables below for 100 nM final stimulations with IGF1 and EGF (note that the solutions are prepared at a higher concentration for final dilution to the required 1X amount once added to the cells in volumes of 500 µl). Make sure to mix all stock solutions prior to final pipetting.
| A | B | C |
|---|---|---|
| Solution: IGF1 (5X = 500 nM) | Soution: IGF1 (1X)-BYL719 (6X - 3000 nM) | |
| LCI | 597 µl | 597.6 µl |
| IGF1 (100 µM) | 3 µl | 0.6 µl |
| BYL719 (1 mM) | 1.8 µl | |
| Final V | 600 µl | 600 µl |
Example table for preparation of IGF1/BYL719 treatment solutions for a final addition of 100 nM IGF1 and 500 nM BYL719. Each treatment required is added in volumes of 500 µl, therefore you can scale the master mix volume according to the number of dishes to treat, then aliquot separately for each treatment. Note that the first IGF1 solution is prepared at 5X stock concentration because it is diluted 5X when adding 500 µl to 2 ml of the medium that is already on the cells. The subsequent IGF1-BYL719 solution is added to 2.5 ml medium that already contains IGF1 but does not have BYL719, therefore the latter is prepared at 6X the required concentration due to a final 6X dilution when 500 µl are added to 2.5 ml.
| A | B | C |
|---|---|---|
| Solution: EGF (5X = 500 nM) | Solution: EGF (1X)-BYL719 (6X - 3000 nM) | |
| LCI | 1196 | 1196 µl |
| EGF (161.3 µM) | 3.7 µl | 0.74 µl |
| BYL719 (1 mM) | 3.6 µl | |
| Final V | 1200 µl | 1200 µl |
As above but for EGF; the higher final volumes ensure that no volume to be pipetted falls below 0.5 µl.
Live-cell imaging and treatment additions
Following 2.5 hours of serum-starvation, bring the cells on the microscope and proceed with setting up which takes around 30 min.
Connect the fluidics system to the dish by exchanging the lids. Make sure that the fluidics system is secured (use white Blu-Tack only) and the dish is clamped in place. Ensure stable CO2 and temperature conditions.


Set-up the microscope with the correct laser and exposure settings (these will be microscope-specific, but we advise careful checking of individual signal intensities to avoid saturation).
For KTR widefield-based imaging, we use the 10X objective on a Nikon TiE2 Unit (with perfect focus).
Select individual positions for imaging, making sure that a cell expresses both the KTR (cytoplasmic/nuclear localisation) and the nuclear marker that will be used for segmentation.
Make sure to only select cells that do not have a saturated signal, yet have a signal that is at least twice as strong as the surrounding background.
Specify the number of time points: we usually image every 6 minutes for 5 or more hours, across up to 24 positions, with a 10X dry objective.
Start imaging and proceed with treatments at the required time points. We usually attach the syringes to the fluidics system in advance and 30s before treatment needs to commence, we pipette the solutions in each syringe, then push it through at the required time point, ensuring that we mix gently 3-4 times by performing several strokes of up to 0.5 ml with the syringe.
NB: Be sure to check on CO2 levels throughout imaging. The cells and their signalling responses are very sensitive to fluctuations in CO2 and pH. This is a common reason for inconsistent result generation. 2 levels throughout imaging. The cells and their signalling responses are very sensitive to fluctuations in CO2 and pH. This is a common reason for inconsistent result generation.
Once imaging has completed, clean the fluidics system, clear the microscope out and proceed with data analysis.
To clean the tubings after each experiment, first flush with 3-4 ml 70 % ethanol (using syringe), then 3-4 ml MilliQ water, then push air through 3-4 times and leave to dry.

