PreTect SEE
Bente Marie Falang
Abstract
PreTect SEE is a CE-IVD kit for the qualitative detection of E6/E7 mRNA from HPV 16, 18 and 45 in a single analysis, identifying women at risk of having or developing cervical pre-cancer or cancer related to the carcinogenic HPV types included.
The kit is based on real-time NASBA technology combining nucleic acid amplification and simultaneous detection with Molecular Beacon probes. This process requires isolated nucleic acids as starting material.An Intrinsic sample control (ISC) is included in order to assess specimen quality and reveal possible factors that may inhibit the amplification, hereby monitoring the entire test process.
NASBA is an enzymatic one-step amplification process that is able to amplify RNA under isothermal conditions (41°C). The presence of genomic dsDNA will not cause false positives, as denaturation of dsDNA does not occur at this low temperature.
Cervical cancer is the most common cancer in women under 35 years of age. The main cause of cervical cancer is the human papilloma virus (HPV). Most women are infected with HPV during their lifetime and as many as 10-30 % of a female population below 35 years of age may be infected.
PreTect SEE is focusing on the HPV types with the highest risk of cervical cancer (HPV 16, 18 and 45). To further increase the specificity, only active oncogene expression from the E6/E7 oncogenes are measured. Published research show that 90 % or more of the cervical cancer cases in younger women are caused by only the three HPV types 16, 18, and 45.

Before start
Specimen collection and preservation:
Liquid based media e.g. PreTect TM, PreservCyt/ThinPrep, SurePath have been validated for the use with the PreTect SEE assay.
Users must follow the manufacturer`s instructions for collecting and storage of cervical cell specimens.
The NASBA process requires isolated nucleic acids as starting material, several protocols may be used.
Completion of the isolation of Nucleic acids (Total nucleic acids or RNA) from the specimens allows for amplification and detection of HPV mRNA E6/E7 types 16, 18, 45 and ISC
Steps
Sample preparation and lysis of material:
Mix each sample thoroughly and pipette 1ml (PreservCyt) to a 10 ml sterile centrifuge tube.
Centrifuge cells for 12 minutes at 2500 rpm (1125g)
Discard the supernatant with a Pasteur-pipette or by vacuum suction
Add 1 ml lysis buffer (PreTect X) to the cells and homogenize by vortexing.
Incubate at room temperature for minimum 10 minutes until isolation procedure starts.
Lysed material may be stored at 2-30°C for 24 hours prior to isolation
Automated Isolation of total nucleic acids (can be done manually using magnetic rack)
Hamilton StarLET and PreTect X kit; based on Booms technology.
1 ml lysed material is added 100µl magnetic silica suspension, incubated at room temperature for 5 min.
3 different Wash steps (Wash buffer 1, 2 and 3) prior to elution.
Elution in 80 µl buffer at 60`C for 5 minutes.
PreTect SEE assay: Amplification and detection of HPV mRNA E6/E7
Bring all kit reagents and samples to room temperature prior to processing.
Allow the prefilled PCR plate to thaw at room temperature for 10 min protected from light.
Centrifuge the PCR plate for 10 seconds at 500g prior to addition of samples.
Please note that positive control is pre-filled into well A1 and negative control into well B1.
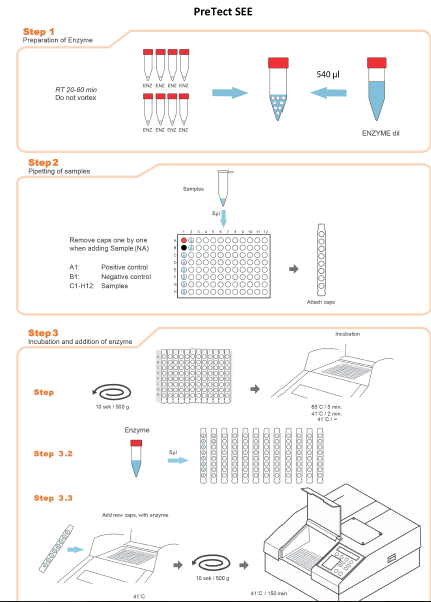
Illustration: Procedure at a glance
Reconstitution of Enzyme
Open the 8 foil packs (ENZ) containing 1 enzyme sphere each and collect all in one tube.
Add 540 ml ENZdil (Enzyme diluent) to the tube containing 8 enzyme spheres.
Leave the enzyme solutions at room temperature for 20-60 minutes until they come to a homogenous solution.
Spin down tubes briefly. Do not vortex.
Addition of samples and preincubation
Transfer 5 µl of each sample (Nucleic Acid) in the positions according the defined plate layout.
Spin the plate (minimum 500g, 10 seconds).
Incubate the 96 well PCR plate for 5 minutes at 65 ± 1 °C and 2 minutes at 41 ± 0.5 °C using a standard PCR cycler.
Addition of enzyme
During incubation step, place 12 new cap strips upside down and add 5µl enzyme solution to the inner side of each cap strip.
After completion of incubation step, remove the cap strips one by one and replace with new cap strips prefilled with enzyme.
Use a Centrifuge with 96 well plate adapter (minimum 500g, 10 seconds) to spin the enzymes directly into each well.
Real time measurement (Analyzer)
Place the PCR-plate immediately into the pre-heated (41ºC) Analyzer and follow the instructions in the PreTect Analysis Software Operator Manual to start, review and accept a run.
Leave the Analyzer running for 150 minutes. The PAS software writes the results to the nc1-file continuously.
PAS supports the following instruments:
· PreTect Analyser (BioTek FLx800 BIE)
· NucliSENS EasyQ® Analyzer (BioMerieux)
· CFX96 (BioRad)
· Quant Studio 5(Applied BioSystems)
PreTect Analysis Software (PAS)
PAS is a dedicated application software package for planning the layout of the PreTect SEE assay setups on a fluorescence reader, retrieve measurement data and interpret, present, and finally report the complete analysis data and sample results.The PreTect Analysis Software performs all assay validation.
For details on how to operate the PreTect Analysis Software, refer to the dedicated PAS Operator Manual.
The PreTect Analysis Software package is available in several languages.
The real-time measurement data are analysed and the final results are presented as shown.
All sample results are presented in detail for both the internal control (ISC) and the HPV types, presented in columns for identification of Sample ID, well positions and result.
Assay Controls/Error messages
PAS checks and validates all positive and negative controls for HPV 16, 18, 45 and ISC.
All controls have to be valid in order to report HPV mRNA results.
If all positive/negative controls are valid a statement is added to the Messages section of the sample report.
Intrinsic sample control
All specimen ISC results are checked and validated by PAS.
ISC monitors the isolation, amplification and detection steps of the assay, revealing possible factors that may inhibit the amplification. The ISC must give a positive result for a negative test result to be valid.
If an ISC is invalid, an error message is given for the relevant samples and an explanation is found in the Messages section of the sample report.
Interpretation of results
Clinical interpretation of the results should be made in conjunction with all other clinical findings for the patient and patient history.
PreTect SEE detects the presence of oncogenic activity and genotype individually HPV types 16, 18 and / or 45. These three HPV types cause more than 90 % of cervical cancer in younger women.
Negative test result
Oncogene activity caused by HPV types 16, 18 or 45 has not been detected.
A negative PreTect SEE test does not exclude the possibility of an existing HPV infection, but merely indicates the absence of detectable HPV oncogene mRNA from HPV types 16, 18 and 45.
Results depend on adequate specimen collection, absence of inhibitors and sufficient mRNA to be detected.
Positive test result
Oncogenic activity of one or more of the HPV types 16, 18 and 45 has been detected.
It leads to increased risk for high grade cervical dysplasia, even if the coinciding Pap smear is normal.
A positive PreTect SEE test result is not a diagnosis of cervical cancer or high-grade cervical lesions.
Governmental instructions and recommendations for the monitoring of HPV status should be followed carefully.
Invalid test result
It has not been possible to obtain a valid test result. Improper specimen collection, storage or specimen processing may have caused the invalidity.
It is recommended repeat isolation or to collect a new sample from the patient, following adequate procedures to ensure high quality and a reliable test result.

