Populating NCBI template for submissions using BioNumerics v7.6
Ruth Timme, Maria Balkey, Julie Haendiges
Disclaimer
Please note that this protocol is public domain, which supersedes the CC-BY license default used by protocols.io.
Abstract
PURPOSE: to define the standard operating procedure for collecting isolate metadata using BioNumerics for submission of food/environmental isolates to NCBI.
SCOPE: to provide a standardized procedure to collect isolate metadata using BioNumerics for submission of food/environmental isolates to NCBI.
RESPONSIBILITIES - SOP Responsible Officials: Ruth Timme, Maria Balkey
The GenomeTrakr Network Management will be responsible to monitor GenomeTrakr submissions processed through Bionumerics and ensure that all GT labs are familiar with the mandatory metadata fields required for submission of GenomeTrakr sequencing records to NCBI.
Steps
Metadata SampleSheet preparation
Before uploading your sequencing run or linking NCBI sequencing records at the BioNumerics platform make sure to fill out the metadata spreadsheet form.
Please download the template and guidelines included in the file 'GT_BioNumerics_spreadsheet_v2.xlsx'.
Create the fields NCBI_bioproject, Attribute_package, Organism_name, NCBI_LabID, SourceCountryState, Latitude_longitude, Reference_material, Culture_collection or Description if they are not in the BioNumerics interface and are needed to process the metadata for your isolates.
Once you have filled out the template information, save the template sheet as .csv and import the metadata to BioNumerics.
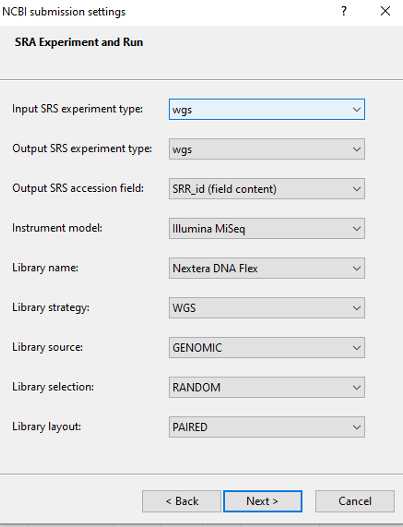
NCBI Submission Settings (Manage submission template)
Create the NCBI metadata template in BioNumerics following PulseNet instructions making sure fields are populated according to GT requirements which are described in the following steps.
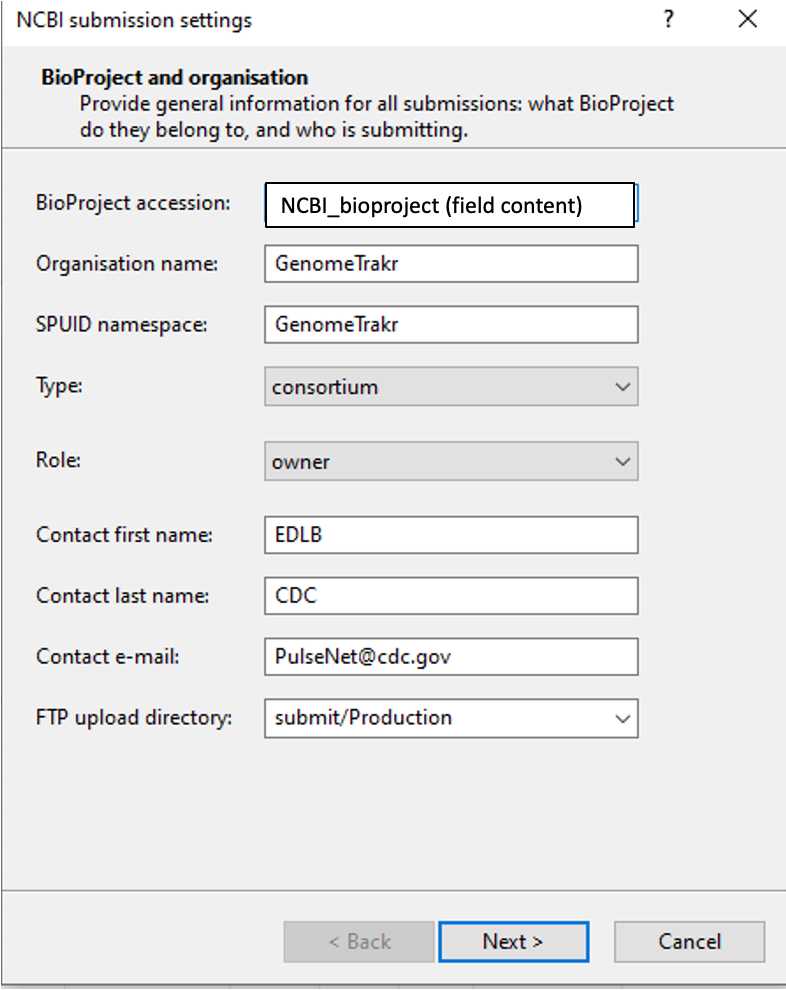
BioProject and Organization: GenomeTrakr labs by submitting independently become owners of their data and are responsible for managing individual bioprojects for each sequenced organism. The term 'field content ' field content ' denotes that the template value e.g. BioProject accession is mapping to the field in BioNumerics e.g. NCBI_bioproject.
| A | B | C |
|---|---|---|
| Name of Field in BioNumerics Template | Description | Example |
| BioProject accession | Identifier for NCBI data collection that contatins data associated to GenomeTrakr. Specific for organism and lab submitter | PRJNA514285 |
| Organization name | Surveillance Program (example is the default value for GenomeTrakr submissions) | GenomeTrakr |
| SPUID namespace | Surveillance Program (example is the default value for GenomeTrakr submissions) | GenomeTrakr |
| Type | organization type (example is the default value for GenomeTrakr submissions) | consortium |
| Role | laboratory responsibility (example is the default value for GenomeTrakr submissions) | owner |
| Contact first name | First name for Lab POC for NCBI submissions. Lab might choose to create alias name for WGS team | First Name |
| Contact last name | Last name Lab POC for NCBI submissions. Lab might choose to create alias name for WGS team | Last Name |
| Contact e-mail | email for Lab POC for NCBI submissions. Lab might choose to create alias name for WGS team | first.last@lab.gov |
| FTP upload directory | Name of directory at NCBI FTP site (example is the default value for GenomeTrakr submissions) | submit/Production |
Table 1. Guidelines for Bioproject and Organization metadata
Laboratories will be submitting to specific bioprojects for lab/organisms. Find the organism/lab specific bioproject under each of the GenomeTrakr umbrella bioprojects included at https://www.ncbi.nlm.nih.gov/bioproject/593772
Make sure to submit to your lab bioproject. Please don't submit to umbrella bioprojects.
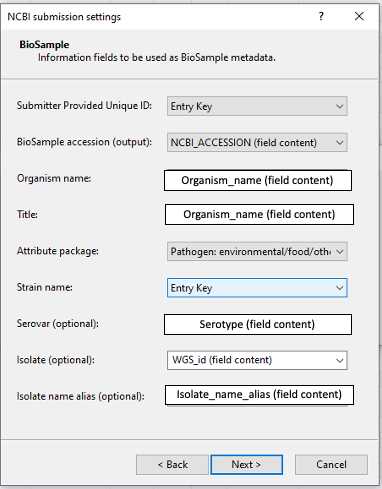
BioSample: Metadata associate to the isolate might require the creation of new fields in BioNumerics. The term 'field content ' field content ' denotes that the template value e.g. Organism name is mapping to the field in BioNumerics e.g. Organism_name. The template values might map to default values e.g. Pathogen: environmental/food/other; version 1.0. Make sure to include the metadata associated to the isolates in the mandatory fields such as: Submitter Provided Unique ID, BioSample accession (output), Organism name, Title, Attribute package, Strain name and Isolate name alias. Isolate name alias is a mandatory field for GenomeTrakr submissions . Provide serovar when available.
| A | B | C | D |
|---|---|---|---|
| Name of Field in BioNumerics Template | Description | Name of Field in BioNumerics DataBase | Example of metadata value |
| Submitter Provided Unique ID | Local lab strain ID | Entry Key | 21B00181-5 |
| BioSample accession (output) | NCBI accession will get populated upon submission to NCBI | NCBI_ACCESSION (field content) | SAMN17385051 |
| Organism name | Genus – species for organism | Organism_name (field content) | Listeria monocytogenes |
| Title | Organism name | Organism_name (field content) | Listeria monocytogenes |
| Attribute package | Sample category | Pathogen: environmental/food/other; version 1.0 | Pathogen: environmental/food/other; version 1.0 |
| Strain name | PNUSA identifier (automatically populates at the time of registration) | WGS_id (field content) | PNUSAL008933 |
| Serovar (optional) | Serotyping information for Escherichia coli and Salmonella enterica | Serovar (field content) | missing |
| Isolate (optional) | Field is not required for GenomeTrakr | missing | |
| Isolate namea alias (optional) | Optional identifier for collaboration projects | Isolate_name_alias (field content) | 21B00181-5; RS_21290 |
Table 2. Guidelines for BioSample metadata
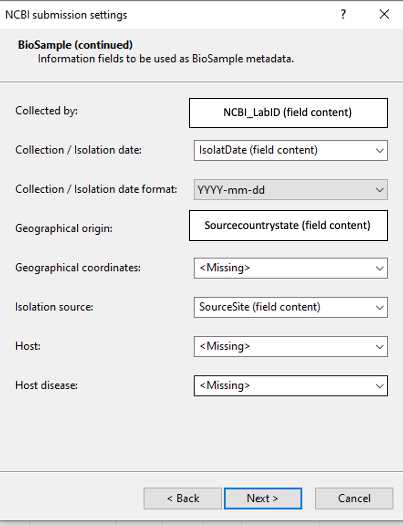
BioSample: Make sure to include the metadata associated to the isolates in the mandatory fields such as: Collected by, Collection / Isolate date, Collection / Isolate date format, Title, Geographical origin and Isolate source. Isolate name alias is a mandatory field for GenomeTrakr submissions. Provide Geographical coordinates when available. Host or host disease are provided only for isolates obtained from human, indicate "missing" for isolates from food or environmental sources.
| A | B | C | D |
|---|---|---|---|
| Name of Field in BioNumerics Template | Description | Name of Field in BioNumerics DataBase | Example of metadata value |
| Collected by | Full name of laboratory that collected the sample or has taken over curation of the isolate. | NCBI_LabID (field content) | NY Department of Agriculture and Markets |
| Collection date | Date on which the sample was collected. | IsolateDate (field content) | 2020 |
| Geographical location | Country and State for sample collection | SourceCountryState (field content) | USA:NY |
| Geographical coordinates | latitude and longitude for site of collection. Missing if it is not provided | missing | |
| Isolation source | Detailed description for sample product or environmental source | SourceSite (field content) | cheese |
| Host | Only provided for human isolates | missing | |
| Host disease | Only provided for human isolates | missing |
Table 3. Guidelines for BioSample metadata (2)
NCBI submission settings – Submission Template
Save submission template according to PulseNet Instructions as - GenomeTrakr-Template -.
Import data
Import the GenomeTrakr Metadata form for BioNumerics ( GT_BioNumerics_spreadsheet_v2 .csv ) according to PulseNet Instructions.
When importing rules, the field source should match destination fields.
In the importing links section, choose the -key- for linking records to database entries.
Proceed with sequencing data import according to PulseNet Instructions.
Submit data to NCBI according to PulseNet Instructions. If NCBI accessions are not available at BioNumerics in 1 business day, please contact NCBI and PulseNet to troubleshoot issues with submissions.
Contact GenomeTrakr by email genometrakr@fda.hhs.gov if issues with submissions are delayed for more than 3 days. GenomeTrakr can support urgent submissions if needed.
NCBI submission for fields not included in the BioNumerics Template.
Laboratories need to include the name of the laboratory sequencing the isolates and the surveillance effort name in the sequence_by and project_name fields, respectively. After receiving biosample accessions, fill out the BioNumerics_update.xlsx spreadsheet and submit the update for these fields to NCBI by contacting biosamplehelp@ncbi.nlm.nih.gov.