Nuclei isolation, immunostaining, and Fluorescence-activated nuclei sorting (FANS) for Smart-Seq2
Iain Macaulay, Ester Kalef-Ezra, Christos Proukakis, Dominic Horner, George Morrow, Yanping Guo, Vanda Knitlhoffer, Andy Goldson
ASAPCRN
Smart-Seq2
scRNA-seq
Sorting
FANS
Brain
Immunostaining
Nuclei Isolation
Aligning Science Across Parkinson’s
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
This protocol describes the steps for extracting nuclei from human postmortem brain samples, immunofluorescence, and nuclei sorting (FANS) for single-nucleus RNA-seq using the Smart-Seq2 method.
We have used it to isolate nuclei from human brain samples (such as the cingulate cortex), but it can be adapted for nuclei from different body areas, cell culture materials, and/or different species.
Steps
Nuclei isolation from human post-mortem brain tissue
Preparation:
UV-treat 96-well plates prior to use.
Pre-weight tubes are needed for tissue scaling.
Clean pestles the day before with 0.2Molarity (M) NAOH, 10% Presept and place them in a falcon tube containing RNase AWAY Decontamination Reagent.
Incubate the pestles with the RNase AWAY O/N @Room temperature.
Prepare PBS (1x, RNase-free) and, Sorting Buffer (1x PBS + 5millimolar (mM)EDTA).
Experimental steps:
Clean Human Tissue handling hood with 0.2Molarity (M) NaOH, 10% Presept, 70% EtOH and dH2O (nuclease-free).
Prepare the Lysis Buffer in the materials section.
Distribute 2µL Lysis Buffer in each well of the plates needed for sorting.
Wash the pestles thoroughly with MiliQ-H2O and then with dH2O (RNase-free) and let them air-dry on a clean wipe tissue before use. Once dried, place them in a clean wipe tissue in a clean container (e.g., falcon tube or sealed bag) and put them in the fridge to pre-cool.
Clean human handling hood with 0.2Molarity (M) NaOH, 10% Precept, 70% EtOH, RNaseZap.
Transfer all materials needed, carefully handle human brain samples in a human handling hood, and wash them with RNaseZap.
Prepare 5% BSA in PBS (1x) and store On ice until use, e.g., for 1 sample.
Add RNasin Plus Ribonuclease Inhibitor to Buffer A and B prior touse to afinal concentration of 0.2 U/µl , e.g., for 1 sample:
- Buffer A:
5µLRNasin Plus Ribonuclease Inhibitor in995µLBuffer A - Buffer B:
5µLRNasin Plus Ribonuclease Inhibitor in995mLBuffer B
Remove tissue from -80°C freezer and transfer it to the human tissue handling hood on dry-ice.
Cut small brain pieces of tissue of interest in pre-weighed 1.5 ml Eppendorf tubes.
Weigh the tissue in a scale aiming for 15mg-30mg of each tissue/donor depending on the downstream needs.
Add 200µL cold Buffer A (containing 0.2 U/µl RNasin Plus Ribonuclease Inhibitor) in each tube containing tissue and place it on wet ice.
Homogenize the tissue using the pestle (pre-chilled) provided by grinding gently with twisting force for 50-60 times.
Add 500µL cold Buffer A (containing 0.2 U/µl RNasin Plus Ribonuclease Inhibitor) to the tube and continue to grind for 20-30 times.
Incubate the tube -20On ice for 0h 5m 0s and carefully transfer homogenate to a filter (column) in collection tube (avoid larger debris that sinks to the bottom of the tube).
Incubate the tube with the cap open at -20°C for 0h 7m 0s.
Cap the filter and immediately centrifuge at 13000x g,4°C.
Centrifuge at 600x g,4°C.
Pour out the supernatant and resuspend the pellet in 200µL PBS with 5% BSA that will be overlaid on top of Buffer B (containing 0.2 U/µl RNasin Plus Ribonuclease Inhibitor) in the next step.
Add 1mL cold Buffer B (containing 0.2 U/µl RNasin Plus Ribonuclease Inhibitor) to a 1.5 ml Eppendorf tube.
Carefully overlay the 200 µl nuclear suspension from step 17 on top of Buffer B by slowly expelling the nuclear suspension against the wall of the tube.
Centrifuge the tube at 1000x g,4°C.
Carefully remove the milky layer by withdrawing it into a 1 ml pipette tip and discarding the rest of the supernatant.
Pour out the remaining Buffer B, leaving 50 µL in the bottom of the ultracentrifuge tube (as it contains the nuclear fraction).
Section 2: Nuclei immunostaining
Prepare Blocking Buffer: 0.8% BSA + 0.2 U/µl RNasin in 10X PBS, pH 7.4 (1x), e.g. for 2 ml:
Resuspend the pellet in 450 µl (or a volume of choice depending on the initial tissue input) of cold Blocking Buffer and resuspend by pipetting up and down gently 5 times.
Separate the samples into 2 tubes each:
a. With Abs: 500 µl
b. Negative Control (with isotype control and/or without antibodies): remaining nuclei supplemented with Blocking Buffer to have a final volume 200 µl.
Incubate the nuclei in Blocking Buffer for 0h 30m 0s @4°C in a rotating wheel or in falcon tubes in a tube roller.
Add antibodies directly to the Blocking Buffer.
Add DAPI (the 1/10.000, e.g. add 5 µl of DAPI diluted 1/100 in PBS 1x) to all nuclei.
Incubate all samples for at least 0h 30m 0s at 4°C in a rotating wheel or falcon tubes in a tube roller.
Carefully discard the supernatant, leaving ∼50 µl of buffer above the pellet.
Resuspended in pre-chilled Blocking Buffer (same volume as before) by gently pipetting up and down 5 times.
Re-pellet nuclei at 800x g,4°C and discard the supernatant.
Wash nuclei again with Blocking Buffer.
Pellet nuclei at 800x g,4°C and carefully discard the supernatant.
Resuspend nuclei in 600 µl (or volume of interest depending on the input tissue) pre-chilled Blocking Buffer and proceed to filtering using Flowmi Cell Strainers in clean 1.5 mL Eppendorf tubes.
Transfer the nuclei On ice to sorting facility on ice and perform the sorting as soon as possible.
Section 3. Nuclei sorting (FANS)
Before single-cell sorting, use Accudrop beads for a test sort to evaluate the plate's position and ensure the sorted cells will be deposited into each well accurately in the middle.
As an extra layer of assessment for accurate sorting, assess plate positioning with colorimetric method.
Add 1mL of dH2O into the vial of powder HRP, and dissolve (this stock is 10x concentrated as compared to working solution).
Make a working solution ( 1mg/mL) by:
To get 2mL:
200µLstock HRP (10mg/mL)1800µLdiH2O- 2 drops of Accudrop beads
Keep in the fridge.
To run the test:
Aliquot 2µL of TMB substrate (fridge in the FACS room) into each well of the test plate.
Sort a single bead into a whole plate (or wells needed).
Once the sort is completed, immediately seal the plate and centrifuge (500x g) and wait 0h 5m 0s-0h 10m 0s and count the number of wells that have turned blue.
Spin the sorting plates and arrange plate orientation.
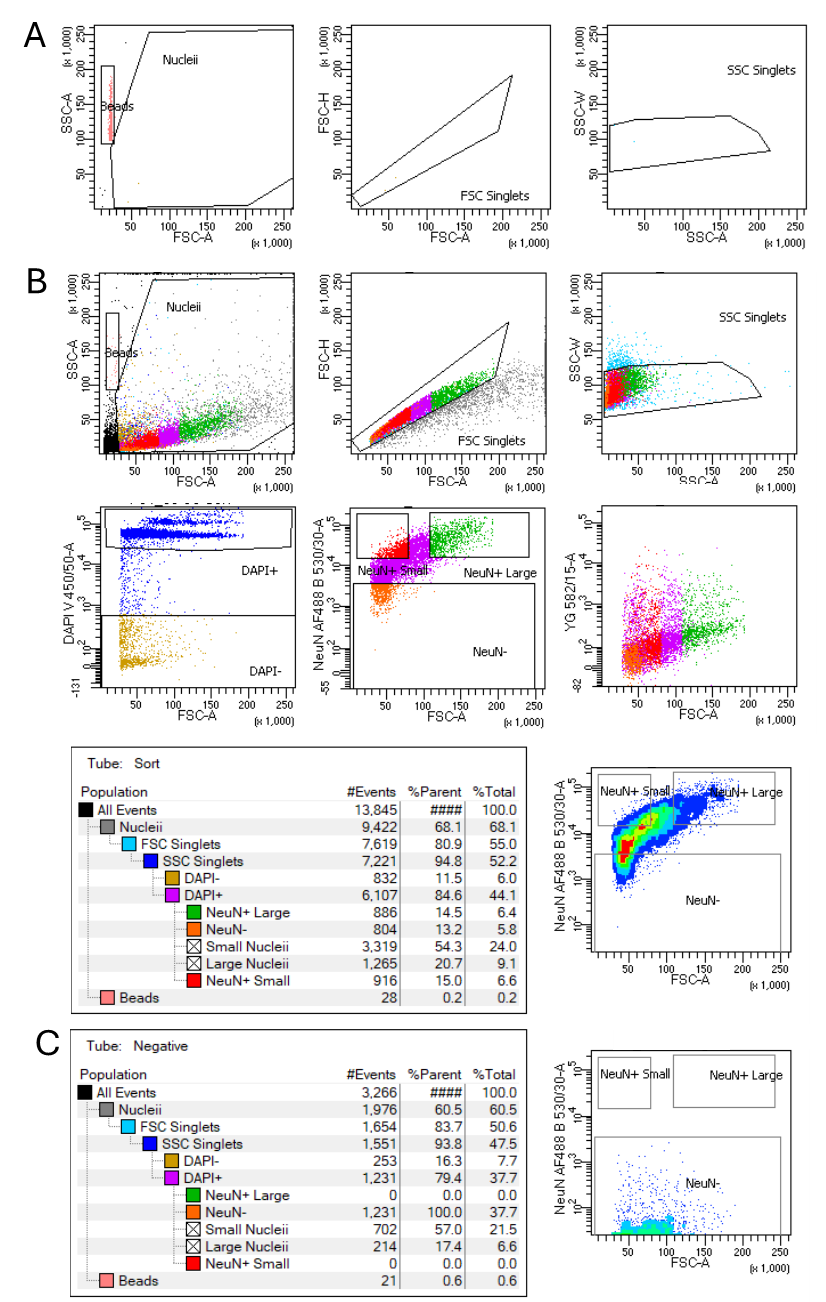
- Select gating parameters to isolate the singlets from the overall detected particles by selecting forward (FSC) and side scatter (SSC), FCS single cell gate, and SSC single cell gate.
- Then select the nuclei by their DAPI expression.
- From the nuclei population (DAPI+), apply further gating parameters based on the antibodies used.
Centrifuge for 0h 0m 10sat low speed the collection plates to ensure reagent at the bottom.
Sort single-nuclei of interest into the centrifuged 96-well collection plates.
After sorting, seal carefully the plates with adhesive plate foil and a seal and place On ice.
Immediately centrifuge briefly at 500x g,4°C.
Place each plate in an individually sealed bag (dry-ice resistant).
Place immediately on dry-ice and transfer to the lab if needed.
Store the plate(s) -70°C until further use and transfer the plates on dry-ice, if necessary, until single-cell Smart-Seq2.
Section 4. Single-nucleus Smart-Seq2
Smart-Seq2 is performed according to Picelli et al. 2014 (Nature Protocols) with the following modifications:
o cDNA is amplified with 25 cycles of PCR.
o oligo-dT30VN, template-switching oligonucleotide (TSO), and IS PCR primers are modified by 5′ biotinylation (Zeisel et al. 2015, Science) (all ordered from IDT).
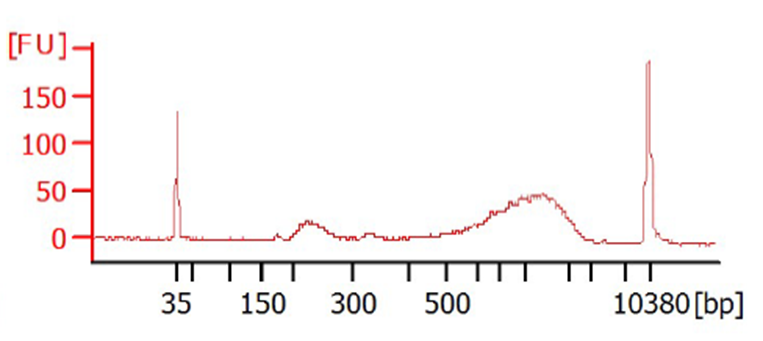
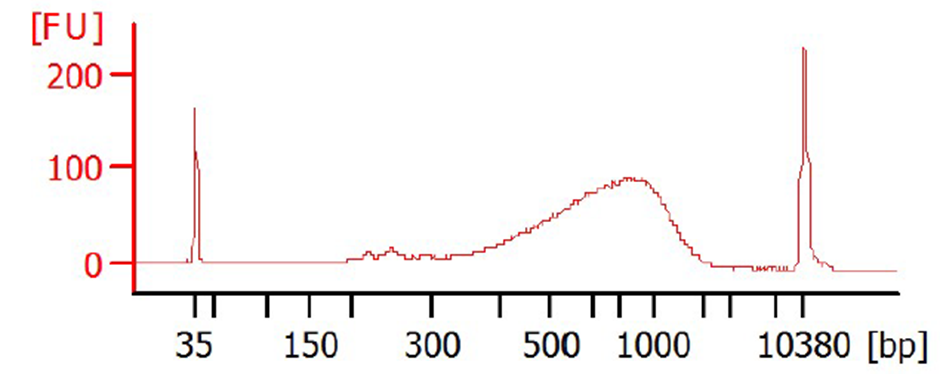
0.6X SPRI-bead clean-up is performed on the cDNA to minimize the presence of short fragments (e.g., primer-dimers) using Biomek NX (Beckman Coulter) liquid handling robot.
cDNA samples are normalized to0.2ng/µL based on an average concentration of 11 samples ran on the bioanalyzer.
Library construction is performed using the Nextera XT sample prep kit (Illumina), a miniaturized protocol involving 12 cycles of PCR.
Libraries are prepared in 384-well PCR plates. The I.DOT (Dispendix) instrument is used to array reagents and indices.
The pooled library is sequenced in an Illumina sequencer.