Nuclei extraction for 10x Genomics Single Cell Multiome ATAC + Gene Expression from frozen tissue using Singulator™ 100 or 200 (S2 Genomics) V2.0
Ronan Chaligne, Ignas Masilionis, Ojasvi Chaudhary, Brigita Meskauskaite Urben
Abstract
This protocol describes nuclei extraction from snap frozen tissue using Singulator, including washing nuclei suspension in buffer supplemented with sucrose, and nuclei FACS sorting. The resulting nuclei suspension is suitable for Single Cell Multiome ATAC + Gene Expression using 10x Chromium platform. The protocol has been validated on various healthy and cancer snap frozen human and mouse tissues (lung, brain, prostate, core needle biopsies etc) .
Before start
Steps
Preparation
-
Turn on and pre-cool Singulator
0h 10m 0s -
Make sure there is sufficient number of Nuclei cartridges (or NIC+ cartridges for samples <20mg) stored / pre-cooled at
4°C
Take the tissue out of LN2 or -80C freezer and place it on dry ice to keep it frozen
Ideally, piece should be <100mg (if not cut it in smaller pieces: <100mg and >10mg).
Cutting can be done with razor blade and within a petri dish on dry ice. Hold the tissue with cooled and RNAse free tweezers (be careful it might fly off). Work fast to not let the tissue thaw while cutting. Place tissue back in a tube on dry ice immediately after cutting.
Prepare 2mL of Nuclei Wash Buffer per sample:
Tris-HCl (pH 7.4): 10millimolar (mM) final
NaCl: 10millimolar (mM) final
MgCl2: 3millimolar (mM) final
Purified BSA: 1% final
DTT: 1millimolar (mM) final
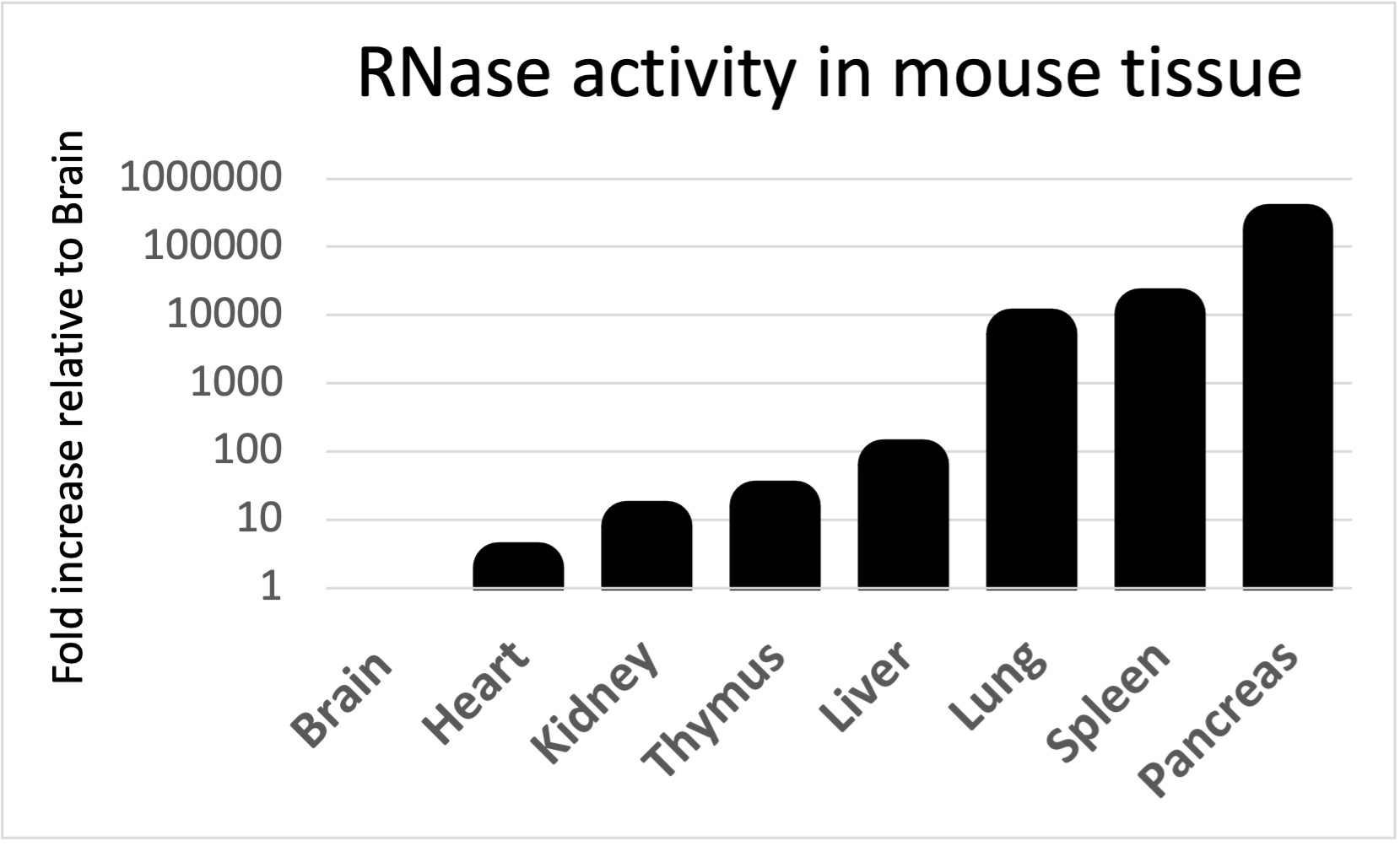
0.2 - 1.0 U/ul RNAse inhibitor (depending on tissue type, see RNase Activity in Mouse Tissue below) (
Top-off to 2mL with Ambion Nuclease-free water
Singulator
Add the following directly into Nuclei Isolation Cartridge before adding your tissue:
RNAse inhibitor (0.2-1.0 U/uL final depending on tissue type - Final volume ~ 3.5 mL for standard nuclei isolation protocol)
DTT, 1mM final concentration (3.5uL of 1Molarity (M) DTT for standard nuclei isolation protocol).
Transfer tissue into the cooled Nuclei Isolation Cartridge and start standard nuclei isolation protocol.
Washes and Prep for FACS
When run is over on Singulator, transfer nuclei suspension (~3.5mL for standard protocol) into 5mL Protein LoBind eppendorf Tubes (or split 3.5mL evenly into four 1.5mL Protein LoBind eppendorf Tubes, conical bottom of the 1.5mL and 5mL tubes increase nuclei recovery).
Add 500uL 2Molarity (M) Sucrose solution to a tube (250millimolar (mM)final conc). Adjust volume if using 1.5mL tubes.
Invert the tube slowly at least 10 times to mix sucrose and nuclei suspension (Do not vortex!).
Make sure sucrose is well mixed, if sucrose cushion is visible after centrifugation, mix again and repeat centrifugation.
500x g,4°C
Nuclei Washes and FACS
After centrifugation, a layer of debris might be visible at the top and a pellet of nuclei at the bottom (for lower yields it might not be noticeable).
Aspirate supernatant with a P1000 pipette, be really careful when removing the debris layer on the meniscus without disrupting nuclei pellet at the bottom of the tube (some liquid can be left to prevent pellet aspiration ~50uL).
Re-suspend Nuclei pellet in ~400uL Nuclei Wash Buffer.
This can be adjusted based on tissue input and pellet size.
OPTIONAL:
A second wash with Nuclei Wash buffer can be performed to remove traces of sucrose (as it might impact FACS sorting).
A second wash with 250mM sucrose can be performed if high amount of debris or fat is still present in the sample.
Filter nuclei suspension through a 35 μm FACS tube cap filter (blue) or 30 μm Pre-separation filters (Miltenyi).
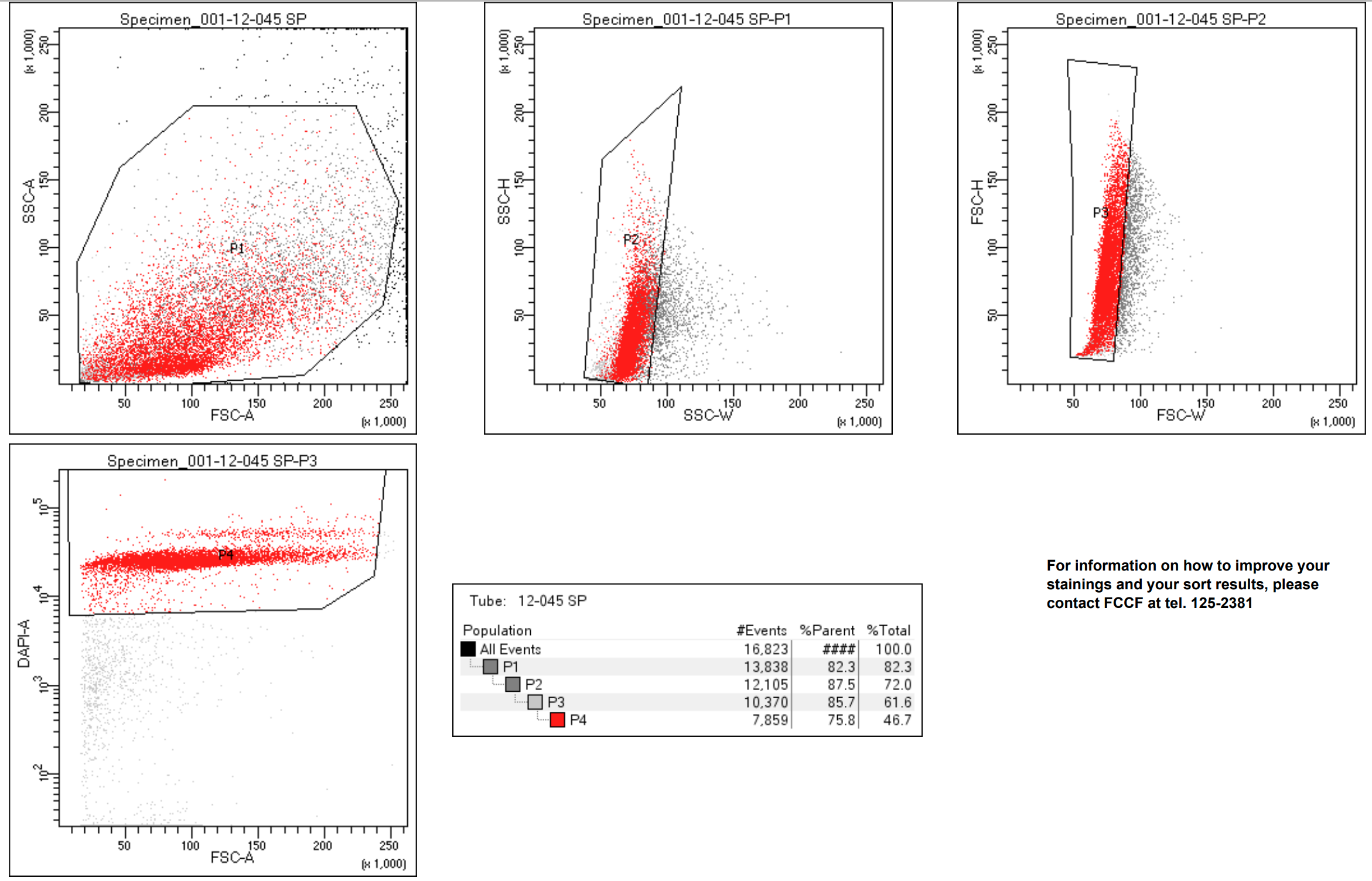
Count nuclei using DAPI on automated cell counter (Countess II for instance)
Trypan can also be used to count in Bright field, but in our experience DAPI count is more accurate.
Nuclei should be stained with
Suggested nozzle size: 100um.
Ideally, aim to collect >200 000 nuclei.
Sort_Report_DAPI_pos_Nuclei.pdf
10x Genomics
Concentrate the nuclei suspension post-sorting:
500x g,4°C
Then, aiming for >=1000 nuclei/uL using FACS numbers, re-suspend sorted nuclei with nuclei Wash buffer .
Count nuclei using DAPI on Countess II cell counter.
Trypan can also be used to count in Bright field, but DAPI counts are more accurate.
Aliquot 200k nuclei in 0.2mL PCR tube.
500x g,4°C
Remove supernatant leaving 5uL precisely (calculate: (Aliquot volume minus 5uL) and aspirate that amount, eyeballing it is highly discouraged) .
Then continue to 10X Genomics Nuclei Isolation for Single Cell Multiome ATAC + Gene Expression Sequencing protocol CG000365. Following " Low Cell Input Nuclei Isolation " in Appendix starting at step e. , omit NP40 from lysis buffer and perform lysis for 30s.

