Nuclei Prep from Frozen Mouse Brain Tissues with Optional Sucrose Column for single nuclei RNA-seq
William F Flynn, Greggory A Perry, Sandra L Daigle, Paul Gabriel, Diane Luo, Jessica Grassmann, Paul Robson, Elise T Courtois
Disclaimer
This protocol has only been tested on a limited number of tissues which includes human and mouse brain frozen tissue. Before using this protocol in production on your tissue of interest, run a pilot experiment to determine optimal dissociation of your tissue.
Abstract
This SOP aims to extract nuclei from unfixed snap-frozen mouse prefrontal cortex, hippocampus, and Striatum tissues (~3-15mg). It was optimized and tested on mice ranging in age groups from 3, 6, 14, and 15-month-old mice on different diets, including high-fat (45%) and regular chow (6%). Other differences included sex, strain, and genotype. These preps resulted in a minimum of debris for downstream snRNAseq HT assays on the 10X Genomics chromium X controller (HT, High Throughput chips).
This protocol was also used to perform snRNAseq assays from human glioblastoma snap frozen tissues. Follow BSL-2 guidelines when working with human tissues. These preps resulted in a minimum of debris for downstream snRNAseq ST assays on the 10X Genomics chromium X controller (ST, Standard Throughput chips).
This protocol enables to process up to 8 samples simultaneously.
Before start
A. Pre-cool centrifuge, buffers, and consumables with sample contact (e.g., gentleMACS C Tube and Strainers) at 4 °C.
B. Per extraction, add 0.2U/ul RNase inhibitor to one 1.6 ml aliquot of Miltenyi Nuclei Extraction Buffer ( 8µl of 40U/uL RNase inhibitor) 8µl of 40U/uL RNase inhibitor ) and keep on ice.
C. Per extraction, add 1 mM DTT to one 750 µl aliquot of Sucrose Solution (0.58µl 1.3M DTT) in a 2 ml tube 0.58µl 1.3M DTT) in a 2 ml tube and keep on ice.
D. Per extraction, prepare 750ul Nuclei Suspension Buffer per sample .
Steps
Procedure- gentleMAC
Add 1250µL On ice to each pre-cooled gentleMACS C Tube.
Transfer frozen tissue piece(s) into the gentleMACS C Tube containing lysis buffer and directly proceed with the following steps until samples are dissociated.
Note: Do not let frozen samples thaw before dissociation, as endogenous RNase might degrade RNA.
Run gentleMACS Program based on tissue size on the gentleMACS Dissociator at 4°C.
See note C under Guidelines & Warnings.
Spin down nuclei, 500x g,4°C in a swinging bucket.
Remove supernatant and resuspend nuclei in 500µL
If the pellet is loose , leave in the tube 50µL in the tube.
See the Guidelines & Warning section D to decide to proceed to step 2 (sucrose column) or 3 (no sucrose column) next.
Sucrose Column
Sucrose Column Purification
In a 2mL microcentrifuge tube with 750µL, layer 500µL on top, forming two layers.
Spindown, 16000x g,4°C in a fixed angle rotor.
Carefully pour off and blot the tube or aspirate the debris with the supernatant. Remove as much supernatant as possible without disturbing the nuclei pellet.
Resuspend the nuclei pellet in 50-150µl Nuclei Suspension Buffer and let sit on ice without pipetting for 3-5 minutes to loosen the pellet.
Pre-wet 40μm filter with Nuclei Suspension Buffer.
Gently pipette mix 10-15x with wide bore tip and filter through pre-wet 40μm filter into new cold 1.5mL tube.
Be careful not to wash down sucrose waste from the tube walls.
Proceed to Step 4 and 5.
Wash without Sucrose Column
Washing nuclei without the use of a sucrose column
Add 500µL and gently pipet mix 5-10x using wide bore tip to suspend nuclei.
Centrifuge in a swinging bucket 350x g,4°C.
Remove supernatant, being careful not to disturb the nuclei pellet.
Repeat Steps 3.1 - 3.3 for a total of 2 washes.
Resuspend the nuclei pellet in 50-150µl Nuclei Suspension Buffer and let sit on ice without pipetting for 3-5 minutes to loosen the pellet.
Pre-wet 40μm filter with Nuclei Suspension Buffer .
Gently pipette mix 10-15x with wide bore tip and filter through a pre-wet 40μm filter into new cold 1.5mL tube.
Proceed to step 4 and 5.
Nuclei counting
Nuclei counting with Trypan Blue / AOPI stain
Obtain nuclei in suspension buffer from steps 2 or 3.
Proceed to Step 4.1 if using Luna FX7 cell counter and AOPI stain to count nuclei.
Proceed to Step 4.2 if using Countess FL II cell counter and Trypan Blue stain to count nuclei.
Proceed to Step 4.3 if using Nexcelom Cellometer K2 and AOPI stain to count nuclei.
Note: Trypan Blue staining is useful to visualize debris and nuclei qualitatively. The counts using Trypan include both nuclei and debris. AOPI is used to count only nuclei. By comparing Trypan and AOPI counts, one can quantitatively asses the amount of debris present in samples preps.
Nuclei counting with LunaFX7
A. Add 1uL AOPI stain and 9uL suspended nuclei to a .5 ml microcentrifuge tube or PCR strip tube and mix gently by pipetting 5x.
Examples:
1ul AOPI/9ul sample
1ul AOPI/4.5ul sample/4.5 ul suspension buffer
B. Turn on Luna FX7 cell counter.
Select "fluorescence cell counting" and "cell lines & primary cells" with the touch screen menu.
C. Select the settings tab and choose the correct slide and wells being counted.
D. Load preferred protocol for fluorescence cell counting.
E. Load preferred protocol for fluorescence cell counting.
F. Select Count, insert the slide into the LunaFX7, and pre Count on the right-hand menu.
G. Record desired data after Count is finished using the save and print command, which allows renaming of the sample, or use quick save, which auto-generated a name.
H. Below is an example of Luna FX7 images.
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.81wgb657ylpk/image%20%281%29.png" alt="Left image shows "dead" cells or nuclei (red, nuclei stained with PI).Right image shows the brightfield for the same field." loading="lazy" title="Left image shows "dead" cells or nuclei (red, nuclei stained with PI).Right image shows the brightfield for the same field."/>
Nuclei counting with Countess FL II
A. Add 6 uL Trypan Blue stain 0.4% and 6 uL nuclei suspension to 0.2 mL PCR strip tube. Mix gently by pipetting 5-10x.
B. Load 10 uL into Countess cell counting chamber slide.
C. Visually assess nuclei sample in slide using bench top microscope, 20x magnification. Note debris present, if any, and overall nuclei shape/integrity.
D. Load slide into Countess FL II. Adjust brightness and focus if necessary. Click "Count".
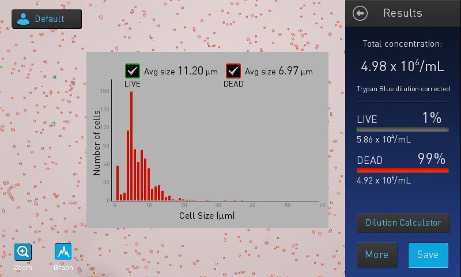
E. Once counting has been completed, record nuclei concentration, viability, and size. Save data with correct date and sample name.
F. Below is an example of Countess FL II readout showing "dead" cells, or nuclei.
Nuclei counting with Nexcelom Cellometer K2
A. Add 11 uL AOPI stain and 11 uL nuclei suspension to 0.2 mL PCR strip tube. Mix gently by pipetting 5-10x.
B. Load 20 uL into Cellometer SD100 slide.
C. Record nuclei concentration, viability, and size.
Nuclei quality QC: Hoechst and SYTO RNASelect staining of nuclei
Hoechst and SYTO RNASelect staining of nuclei
Keep all stains and dilutions on ice in the dark 4°C.
Choose 5.1 for Leica DM4 B or 5.2 for PerkinElmer Opera Phenix.
Nuclei QC with Leica DM4 B
Imaging:
Microscope - Leica DM4 B
Camera - Leica DCF7000 GT
Exposure time:
Hoechst - 45 ms, gain 5.5
SYTO RNASelect - 800 ms, gain 3.4
Brightfield - 30 ms, gain 8.8
Magnification - 40X
- The exposure time and gain may need to be adjusted depending on the microscope used.
A. Make a 1:80 dilution of Hoechst stain (1ul into 79ul PBS)
B. Make a 1:500 dilution of SYTO RNASelect stain (1ul into 499ul PBS)
C. Mix well and aliquot 18ul of nuclei suspension into a new microcentrifuge tube. Dilute if needed at this point, but the total volume should be 18ul.
D. Add 1ul 1:80 Hoechst stain and 1ul 1:500 SYTO RNASelect stain to 18ul suspended nuclei and mix gently 5x pipetting. Immediately place the mixture in a light-blocking box and place it at ,4°C.
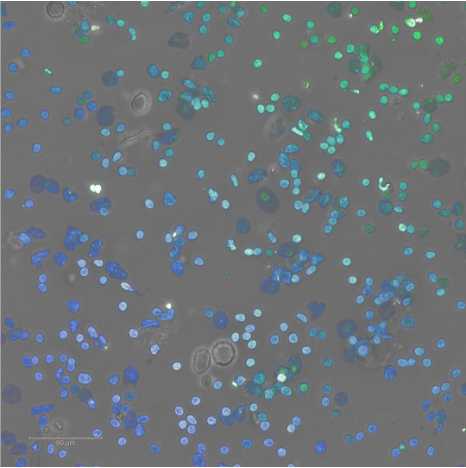
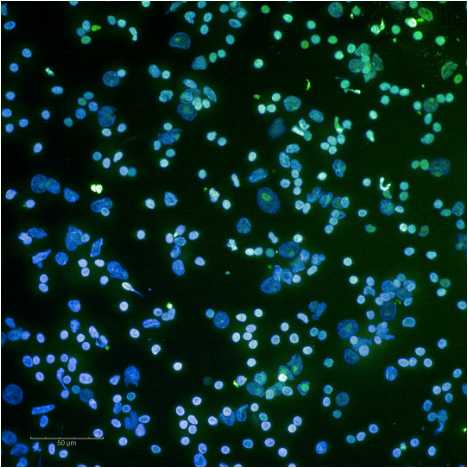
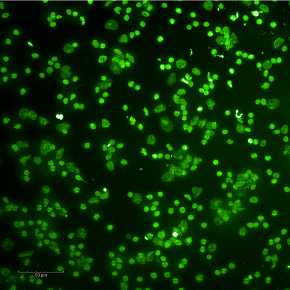
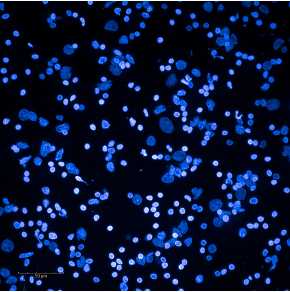
Expected results of nuclei put through a sucrose column are shown below.
See guidelines for further details.

Nuclei QC with Opera Phenix confocal microscope with 40x water objective.
A. Dilute SYTO RNASelect stain 1:1000 and dilute Hoechst 1:100 in DPBS. Keep dye dilutions on ice and protected from light.
B. Place a 384-well plate (PerkinElmer CellCarrier Ultra) on ice.
C. Add 20 uL of nuclei suspension to a 0.2 mL strip tube.
D. Add 1 uL each of diluted SYTO RNASelect and Hoechst stains into tube containing nuclei suspension. Pipet mix 5-10x.
E. Transfer contents of strip tube into 384-well plate. Incubate 384-well plate at ,4°C protected from light.
F. Image 384-well plate using the following parameters, ensuring that no channels are over/underexposed:
**Plate Type:** 384 PerkinElmer CellCarrier Ultra
**Autofocus:** Two Peak (Default)
**Objective:** 40x Water, NA 1.1
**Opt. Mode:** Confocal
**Binning:** 2
**Channel Selection:**
**Brightfield**
Exposure Time = 200 ms
Power = 100%
**SYTO RNA Select**
Exposure Time = 100 ms
Power = 50%
**HOECHST 33342**
Exposure Time = 100 ms
Power = 50%
**Layout Selection (Well)**
Number of Fields = 49
Overlap = 5%
**Layout Selection (Stack)**
First Plate = -1.0 um
Number of Planes = 6
Distance = 3.0 um
Last Plane = 14.0 um
Overall Height = 15.0 um
G. Example nuclei QC images are below.
Loading nuclei on 10x Genomics chip
10x Genomics downstream applications
Follow 10x recommendations for your specific needs.
For a 10x standard chip (ST) and High Throughput (HT) chip, calculate the number of nuclei that needs to be loaded for each channel, and according to the experimental design.
Loading examples:
ST - Targeting 6,000 nuclei
- load 12,000 to recover 12,000 taking into account a 50% capture rate.
HT - Targeting 20,000 nuclei
- load 33,300 to recover 20,000 taking into account a 60% capture rate.
For more detailed guidelines visit [https://www.10xgenomics.com/](https://www.10xgenomics.com) for more details.