Nova-ST Spatial Transcriptomics protocol
Suresh Poovathingal, Stein Aerts, Kristofer Davie
Abstract
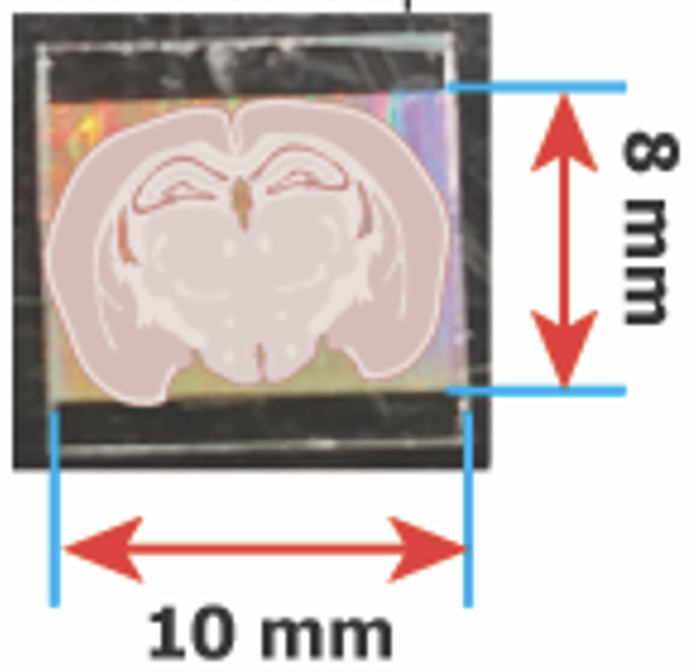
Nova-ST is a an open-source, high-resolution sequencing based spatial transcriptomics workflow. This method gives comparable resolution to BGI Stereoseq, SeqScope & PIXEL seq. Nova-ST is derived from dense nano-patterned randomly barcoded Illumina NovaSeq 6000 S4 sequencing flow cells. More details in the Nova-ST pre-print. Nova-ST enables customized, low cost, flexible, and high-resolution spatial profiling of broad range of tissue section sizes (upto 10mm x 8 mm). In this protocol, we provide detailed step-by-step resource for implementing the Nova-ST spatial transcriptomics workflow in you lab. Bioinformatics and data analysis workflows are detailed in: https://github.com/aertslab/Nova-ST. For any protocol related or data analysis clarifications, you can reach out to us via nova.st.aertslab@gmail.com.
Steps
Tissue preparation for spatial transcriptomics profiling
The first steps of the Nova-ST workflow starts with preparation of tissue for cryo-embedding. Follow the tissue preparation guidelines from 10X Genomics Visium Tissue Preparation guide
Other useful video resources available from 10X Genomics:
Assess the RNA quality of the OCT embedded tissue using the guidelines in the 10X Genomics Visium Tissue Preparation guide. Refer the section: RNA Quality Assessment .
Assessment of tissue permeabilization time
Follow the 10X Genomics Visium Tissue Preparation guide, to place 8 consecutive tissue section in the capture area slots of the Visium Spatial Tissue Optimization Slide (PN: 3000394). The optimization slide with sectioned tissue can be preserved at -80oC as recommended by the manufacturer.
Perform H&E staining of the tissue sections on the Visium Spatial Tissue Optimization Slide using Methanol Fixation, H&E Staining & Imaging for Visium Spatial Protocols.
To perform tissue permeabilization: the protocol in Visium Spatial Tissue Optimization Reagents Kits User Guide is followed with the following key differences:
Prepare 100mg/ml of pepsin stock by dissolving the pepsin powder with 0.1N HCl (2 ). Mix the solution with P1000 pipette, till the enzyme is homogenized. DONOT vortex the enzyme stock. Prepare aliquots of the enzyme stock for single use and store them in -20°C for long term storage. On the day of the experiment thaw the 100mg/ml of pepsin stock on ice.
Prepare 0.65U/ul working stock of Pepsin enzyme by diluting the 100 mg/ml enzyme stock with 0.1N HCl (2 ) warmed to 37°C.
In the Visium Spatial Tissue Optimization Reagents Kits User Guide replace the permeabilization enzyme (PN: 2000214) form the 10X Genomics Visium Spatial Tissue Optimization kit with the pepsin permeabilization reagent prepared above.
Choose a time course with 8 different time point to perform the tissue permeabilization experiment. The time points for the time course may be adjusted depending on the tissue types being handled.
Preparation for Nova-ST profiling
Preparation of Nova-ST chips for Spatial Transcriptomics:
Things to prepare:
- Put the PCR block at 37C and place the 10X Genomics thermocycler adapter (PN:3000380) into the block and let the adapter equilibrate to 37C.
Retrieve the 24-well plate with Nova-ST chip stored in the 1X IDT TE (8 ) buffer from the 4°C fridge. Remove the parafilm seal.
Chose the desired chip number/s from the storage plate. Register the corresponding Nova-ST chip ID/s (this will be needed for later ST data analysis).
Remove and discard the storage TE buffer from the corresponding well/s. Using a sharp forceps retrieve the Nova-ST chip/s and place it into a fresh 24 well plate (the functional surface of the chip must face the top). Ensure not to disturb the functional surface of the Nova-ST while handling the chip
Add 3 ml of NFW, drop by drop on to the surface of the chip to to wash the the chip. Discard the wash and repeat this step for a total of 3X times.
Transfer the chip to fresh well in the 24 well plate. Using either an air gun or pressurized air canister, blow away excess liquid on the surface of the chips. With the help of forceps, transfer the chip to the thermocycler adapter at 37*C. Incubate for 1 min for the chip to dry. Then proceed for the cryo-sectioning of the tissue.
Tissue sectioning for Nova-ST chips:
- Follow the general guidelines and good practices for sectioning of a
10 µmtissue section. Follow the guidelines and recommendations in 10X Genomics Visium Tissue Preparation guide.
Additional useful resources in: Cryosectioning of OCT Embedded Tissue Block.
Label a 3cm petridish with the chip number and couple of glass slides and place them into cryostat.
Allow them to cool to cryostat temperature for atleast 30 mins.
Once the glass slide has cooled to cryostat temperature, add 2 drops of OCT on the glass slide to hold the Nova-ST chip on the glass slide and to prevent the chip from sliding away from the slide.
Place the Nova-ST chip on the glass slide cooled inside the cryostat for 0h 3m 0s prior to sectioning.
Place the sectioned tissue and adjust the position of the tissue to ensure the desired region of interest falls within the functional area of the Nova-ST chip as shown below:
Once the tissue section is positioned correctly on the Nova-ST chip, lift the glass slide and place a finger on the slide underneath where the chip and tissue are located. The tissue melts over the chip surface. Once the chip is completely thawed on the surface of the Nova-ST chip, place the chip back into the cryostat to freeze the tissue. Using a sharp forcep place the chip into the pre-cooled 3 cms petridish inside the cryostat. Perform this gently and carefully to ensure not to disturb the functional surface of the Nova-ST while handling the chip.
If proceeding with spatial transcriptomics workflow proceed with step 9, else transfer the chip in the 3 cm petridish to dry ice and transfer to -80°C for storage. This tissue can be stored at-80°C for 3 weeks.
Tissue hematoxylin and eosin staining & permeabilization
If the chip with the tissue is stored at-80°C , remove the chip and place it on dry ice till the chip is ready for downstream processing.
Preparation for H&E staining:
Things to prepare:
- Put the PCR block at 37C and place the 10X Genomics thermocycler adapter (PN:3000380) into the block and let the adapter equilibrate to 37C.
- Thaw the 100mg/ml of pepsin stock on ice.
- Put two ovens at
37°C(permeabilization) &42°C(first strand synthesis). - Prewarm 0.1N HCl to
37°C. - Thaw dNTP Mix
| A | B |
|---|---|
| Eosin Mix | Volume |
| Eosin Y Solution | 100 ul |
| Tris-Acetic Acid Buffer (0.45 M, pH 6.0) | 900 ul |
| Total | 1000 ul |
- Thaw 1x Maxima H- RT buffer.
- Transfer 12 ml Methanol into a 6 cm petridish (per sample) and place the petridish into
-20°Cfor atleast 15 mins.
Using a sharp forceps transfer the Nova-ST chip/s to the thermocycler adapter at 37°C (without closing the lid of the PCR block). Ensure not to disturb the functional surface of the Nova-ST while handling the chip. Incubate for 2 mins to thaw the tissue on the Nova-ST chip.
After the incubation, transfer the chip into the methanol at -20°C. Perform the methanol fixation for 0h 30m 0s.
During the methanol fixation, prepare the following for the H&E staining:
- Fill 3X 1 L beakers with Milli-Q water
- Prepare 1X 50 ml falcon with Milli-Q water (per sample)
- Prepare Eosin Mix. | A | B | | --- | --- | | Eosin Mix | Volume | | Eosin Y Solution | 100 ul | | Tris-Acetic Acid Buffer (0.45 M, pH 6.0) | 900 ul | | Total | 1000 ul |
After 0h 30m 0s fixation in methanol, using a sharp forceps, transfer the Nova-ST chip from methanol to paper towel to remove methanol from bottom of the chip.
Add 150µL of 2-Propanol to the chip. Ensure the chip is completely covered in the liquid. Incubate for 0h 1m 0s
After the incubation, decant out 2-Propanol and by means of the sharp forcep, transfer the Nova-ST chip to fresh paper towel. Allow the 2-Propanol on the Nova-ST chip to dry-out for 0h 3m 0s . Make sure the chip is completely dry. DONOT exceed 0h 5m 0s.
Perform the rest of the H&E staining of the tissue sections on the Nova-ST using Methanol Fixation, H&E Staining & Imaging for Visium Spatial Protocols.
Some changes to be noted:
- All the volumes of the reagents to be added to Nova-ST chip is
130µL - For wash of the chip with the Milli-Q water, make sure to hold the chip firmly in the region of the chip which doesn't have tissue on it and submerge into water. If the chip is not held firmly, the chip can slip away and get lost.
- For imaging of Nova-ST chip, after drying the stained Nova-ST chip, place the chip on microscopic glass slide, and proceed with imaging.
- During imaging make sure the identify and note down the Nova-ST chip label.
- Note the time for imaging, prior to permeabilization step. DONOT to exceed
1h 0m 0s.
Tissue permeabilization and First Strand Synthesis
Preparations:
Prepare the following mixes (excluding the items marked with **, these items are added just before addition to Nova-ST chip) and incubate for at least 0h 15m 0s in a 37°C oven.
-
Prepare RT wash buffer below (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | Maxima RT buffer | 5 | X | 1 | 80 | | **RNAse Inhibitor (Lucigen) | 40 | U/ul | 0.5 | 5 | | NFW | | | | 315 | | | | | | 400 |
-
Prepare 0.1X SSC below (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | SSC | 20 | X | 0.1 | 2 | | **RNAse Inhibitor (Lucigen) | 40 | U/ul | 0.5 | 5 | | NFW | | | | 393 | | | | | | 400 |
-
Prepare RT First Strand Mix (per reaction):
| A | B | C | D | E |
|---|---|---|---|---|
| Stock | Unit | Final | Volume | |
| Maxima RT buffer | 5 | X | 1.05 | 84 |
| **RNAse Inhibitor (Lucigen) | 40 | U/ul | 1.05 | 10.5 |
| Ficoll PM-400 | 20 | % | 4.5 | 90 |
| **dNTPs (Thermo) | 25 | mM | 1.05 | 16.8 |
| **Maxima H- Rtase | 200 | 10 | 20 | |
| NFW | 178.7 | |||
| 400 |
Prepare 0.65U/ul working stock of Pepsin enzyme by diluting the 100 mg/ml enzyme stock with 0.1N HCl (2 ) warmed to 37°C. Check the pH of the diluted enzyme mix and ensure 2 .
With the help of a sharp forceps transfer the Nova-ST chip after imaging to a 3 cm petridish and add the 120µLpre-warmed 0.65U/ul Pepsin mix onto the chip (add drop-by-drop outside the tissue area). Ensure to spread the liquid evenly on the surface of the Nova-ST chip. Incubate the chip with the enzyme for the optimized permeabilization time in the 37°C oven. During the incubation time, add the remaining reagents in the table (**).
After the incubation time, immediately remove the enzyme from the surface of the Nova-ST chip with repeated aspiration using a P200 pipette. Ensure not to disturb or scratch the functional surface of the chip.
With the help of sharp forceps, transfer the Nova-ST chip to a clean paper towel, to remove the liquid at the bottom of the chip and then transfer the chip to a fresh 24 well plate with the tissue facing up.
Add 0.1X SSC buffer, drop wise, to the corners of the chip to wash the chip. Remove and discard 0.1X SSC buffer. Repeat this step with the 1X RT wash buffer.
Add 1X RT first strand mix, drop wise, to the corners of the chip in the 24 well plate. Ensure that the chip is completely submerged in the liquid. Add 1 ml of NFW to the neighboring wells. Cut 4 square patches of paraflim M. Place the patch on the wells to seal the well with Nova-ST chip. Seal the 24 well plate with parafilm tape and proceed with the incubation to complete the first strand synthesis:
42°C for 16h 0m 0s-20h 0m 0s.
Exonuclease treatment
Preparations:
Things to prepare:
- Put an oven at
37°C. - Thaw 10X Exonuclease buffer
- Thaw NEB buffer 2
- Thaw 25 mM dNTP mix.
- RPE Randomer
100micromolar (µM)(Refer Table S1) - Thaw KAPA HiFi HotStart on ice
- Thaw the RPEPCR*Forward primer
100micromolar (µM)(Refer Table S1) - Thaw the RPEPCR*Reverse primer
100micromolar (µM)(Refer Table S1)
Prepare the following mixes (excluding the items marked with **, these items are added just before addition to Nova-ST chip) and incubate for at least 0h 15m 0s in a 37°C oven.
-
Prepare 0.1X SSC below (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | SSC | 20 | X | 0.1 | 5 | | NFW | | | | 995 | | | | | | 1000 |
-
Prepare 1X Exo-I buffer (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | Exo I buffer | 10 | X | 1 | 40 | | NFW | | | | 360 | | | | | | 400 |
-
Prepare Exo-I reaction Mix (per reaction):
| A | B | C | D | E |
|---|---|---|---|---|
| Stock | Unit | Final | Volume | |
| ** Exo I | 20 | 1 | 20 | |
| Exo I buffer | 10 | X | 1 | 40 |
| NFW | 340 | |||
| 400 |
Prior to adding the mixes to the 24 well plate, add the remaining reagents in (**) to the respective tubes.
After the completion of the FSS step, remove the FSS reagents from the 24 well plate.
Add 0.1X SSC buffer, drop wise, to the corners of the chip to wash the chip. Remove and discard 0.1X SSC buffer. Repeat this step with the 1X Exo-I buffer.
Add Exo-I reaction Mix, drop wise, to the corners of the chip in the 24 well plate. Ensure that the chip is completely submerged in the liquid. Seal the 24 well plate with parafilm tape and proceed with the incubation to complete the exonuclease reaction:
37°C for 0h 45m 0s.
During the exonuclease incubation, prepare the following tissue removal mix (excluding the items marked with **, these items are added just before addition to Nova-ST chip) and incubate for at least 0h 15m 0s in a 37°C oven.
- Prepare 1X Tissue removal mix (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | Tris pH 8.0 | 1000 | mM | 100 | 40 | | NaCl | 5000 | uM | 200 | 16 | | SDS | 10 | % | 2 | 80 | | EDTA | 500 | mM | 5 | 4 | | **Proteinase K | 800 | mU/ul | 16 | 8 | | NFW | | | | 252 | | | | | | 400 |
Tissue removal step
After the completion of exonuclease reaction step, remove the Exo-I reagents from the 24 well plate. Remove as much liquid as possible.
Add pre-warmed 1X Tissue removal mix, drop wise, to the corners of the chip in the 24 well plate. Ensure that the chip is completely submerged in the liquid. Seal the 24 well plate with parafilm tape and proceed with the incubation to complete the tissue removal reaction:
37°C for 1h 0m 0s.
During the tissue removal incubation, prepare the following denaturation and neutralization mixes:
-
Prepare 0.1N NaOH below (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | NaOH | 10 | N | 0.1 | 100 | | NFW | | | | 9900 | | | | | | 10000 |
-
Prepare 100 mM Tris pH 7.5 (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | Tris pH 7.5 | 1000 | mM | 100 | 1000 | | NFW | | | | 9000 | | | | | | 10000 |
First strand denaturation & Second strand synthesis
During the tissue removal incubation step prepare the following second strand synthesis mix (excluding the items marked with **, these items are added just before addition to Nova-ST chip) and incubate for at least 0h 15m 0s in a 37°C oven.
- Prepare 1X Second Strand Synthesis mix (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | NEB Buffer 2 | 10 | X | 1.1 | 44 | | RPE Randomer | 100 | uM | 11 | 44 | | ** dNTPs (Thermo) | 25 | mM | 1.1 | 17.6 | | **Klenow exo (-) Fragment | 5 | U/ul | 0.55 | 44 | | NFW | | | | 250.4 | | | | | | 400 |
After the completion of the tissue removal reaction step, remove the tissue removal reagents from the 24 well plate. Remove as much liquid as possible.
Add 3 ml of NFW drop-by-drop onto the surface of the Nova-ST chip. Remove and discard the wash. Repeat this step for a total of 3X times
Add 3 ml of 0.1N NaOH drop-by-drop to the Nova-ST chip. For completion of the denaturation incubate the chip for 0h 5m 0s, Remove and discard the wash. Repeat this step for a total of 3X times
Add 3 ml of 100 mM Tris pH 7.5 drop-by-drop onto the surface of the Nova-ST chip. Remove and discard the wash. Repeat this step for a total of 3X times.
Finally, add 3 ml of NFW drop-by-drop onto the surface of the Nova-ST chip. Remove and discard the wash. Repeat this step for a total of 3X times. Proceed immediately with the second strand synthesis.
After the completion of last wash of the Nova-ST chip with NFW, with the help of sharp forcep, transfer the Nova-ST chip to a clean paper towel, to remove the liquid at the bottom of the chip and then transfer the chip to a fresh 24 well plate with the tissue facing up.
Add pre-warmed second strand synthesis mix, drop wise, to the corners of the chip in the 24 well plate. Ensure that the chip is completely submerged in the liquid. Cut 4 square patches of paraflim M. Place the patch on the wells to seal the well with Nova-ST chip. Seal the 24 well plate with parafilm tape and proceed with the incubation to complete the second strand synthesis:
37°C for 2h 0m 0s.
During the second strand synthesis incubation prepare the following denaturation mixes:
- Prepare 0.1N NaOH below (per reaction): | A | B | C | D | E | | --- | --- | --- | --- | --- | | | Stock | Unit | Final | Volume | | NaOH | 10 | N | 0.1 | 50 | | NFW | | | | 4950 | | | | | | 5000 |
After the completion of the second strand synthesis reaction step, remove the SSS reagents from the 24 well plate. Remove as much liquid as possible.
After the completion of SSS reaction step, with the help of a sharp forcep, transfer the Nova-ST chip to a clean paper towel, to remove the liquid at the bottom of the chip and then transfer the chip to a 3 cm petridish and proceed immediately with the random primer extension product from the the second strand synthesis. Ensure not the disturb the functional surface of the Nova-ST chip, while handling with the forceps
Add 90µL of 0.1N NaOH, drop-by-drop to corners of the Nova-ST chip. Ensure not to spill out the 0.1N NaOH outside the Nova-ST chip. With a low bind pipette set to 70µL , pipette mix the denaturant on the chip 5X times and incubate for 0h 5m 0s. After incubation, by repeated aspiration with a low bind pipette, remove as much liquid as possible from the surface of the Nova-ST chip and transfer the denatured RPE extract to a low bind 1.5 ml eppendorf tube. (The petridish can be tilted to one of the corners of the chip to facilitate the withdrawal of the RPE product) Ensure not the disturb the functional surface of the Nova-ST chip while with-drawing the RPE extract from the surface of the Nova-ST chip.
Repeat the above step 41 , two additional time for a total of 3X times. Collect all the RPE extract to the same 1.5 ml eppendorf lo-bind tube. To maximize the RPE collect, after the final wash, transfer the chip into a 50 ml falcon and spin the tube at for 500x g . Extract the RPE extract and collect it into the same low bind 1.5 ml eppendorf tube.
Estimate the volume of the RPE extract collected in the denaturation steps above. This volume usually amounts to 250µL . Neutralize the RPE extract by adding 70µL of 1M Tris 7.0. Vortex the content in the 1.5 ml eppendorf tube to mix the content. Incubate for 0h 1m 0s. (Scale the volumes accordingly)
Perform Ampure XP purification as below:
Important: Ensure to vortex and mix the Ampure XP beads well before adding for purification.
-
To purify cDNA add 1.8:1 ratio of Ampure XP beads to sample (
576µLof Ampure XP), and mix by vortexing. -
Incubate at
Room temperaturefor0h 10m 0s. -
Spin down and place on magnet and allow beads to be capture on the magnet. Place on magnet to separate the beads for
0h 8m 0s -
Discard supernatant, and wash once with
1500µLof of freshly prepared 80% ethanol . After0h 0m 30sremove and discard the 80% ethanol. -
Repeat step 4 , 1X additional time.
-
Allow the bead pellet to dry for
0h 10m 0s. Intermittently remove the 80% ethanol flowing down from the bead pellet to facilitate the bead pellet drying. -
Elute cDNA in
44.5µLof elution buffer. Elute out42µLof cDNA library for RPE cDNA product to a fresh PCR tube.
Random Primer Extension product amplification
Prepare PCR mix for RPE PCR:
Note: The KAPA HiFI DNA polymerase with hot start has very low 3-5' exonuclease activity. Still it is recommended to prepare the PCR mix on ice.
| A | B | C | D | E |
|---|---|---|---|---|
| Stock | Unit | Final | Volume | |
| KAPA HiFi Hotstart ready mix | 2 | X | 1 | 50 |
| RPEPCR*F primer | 100 | uM | 1 | 1 |
| RPEPCR*R primer | 100 | mM | 1 | 1 |
| Purified RPE product | 42 | |||
| dH2O | 6 | |||
| 100 |
Prepare the PCR mix above and transfer 58µL of PCR master mix to the PCR tube having the eluted RPE product.
Quick centrifugation to collect reaction to the bottom of the strip tube, before running the following PCR program in a thermocycler for RPE PCR amplification.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial Denaturation | 95*C | 3 minutes | 1x |
| Denaturation | 95*C | 30 seconds | |
| Annealing | 60*C | 1 minute | 14 cycles |
| Elongation | 72*C | 1 minute | |
| Final Elongation | 72*C | 5 minutes | 1x |
| Hold | 12*C | Hold | 1X |
Perform Ampure XP purification as below:
Important: Ensure to vortex and mix the Ampure XP beads well before adding for purification.
-
To purify cDNA add 0.8:1 ratio of Ampure XP beads to sample (
80µLof Ampure XP), and mix by gently pipetting using P200. -
Incubate at
Room temperaturefor0h 5m 0s. -
Spin down and place on magnet and allow beads to be capture on the magnet. Place on magnet to separate the beads for
0h 4m 0s -
Discard supernatant, and wash once with
200µLof of freshly prepared 80% ethanol . After0h 0m 30sremove and discard the 80% ethanol. -
Repeat step 4 , 1X additional time.
-
Spin down the tube to bring down the beads to the bottom of the tube. Place on the magnet to remove the residual 80% ethanol, without disturbing the bead pellet.
-
Allow the bead pellet to dry for
0h 2m 0s. -
Elute cDNA in
25.5µLof elution buffer. Elute out25µLof RPE PCR to a fresh lo-bind 1.5 ml eppendorf tube.
Quality control check
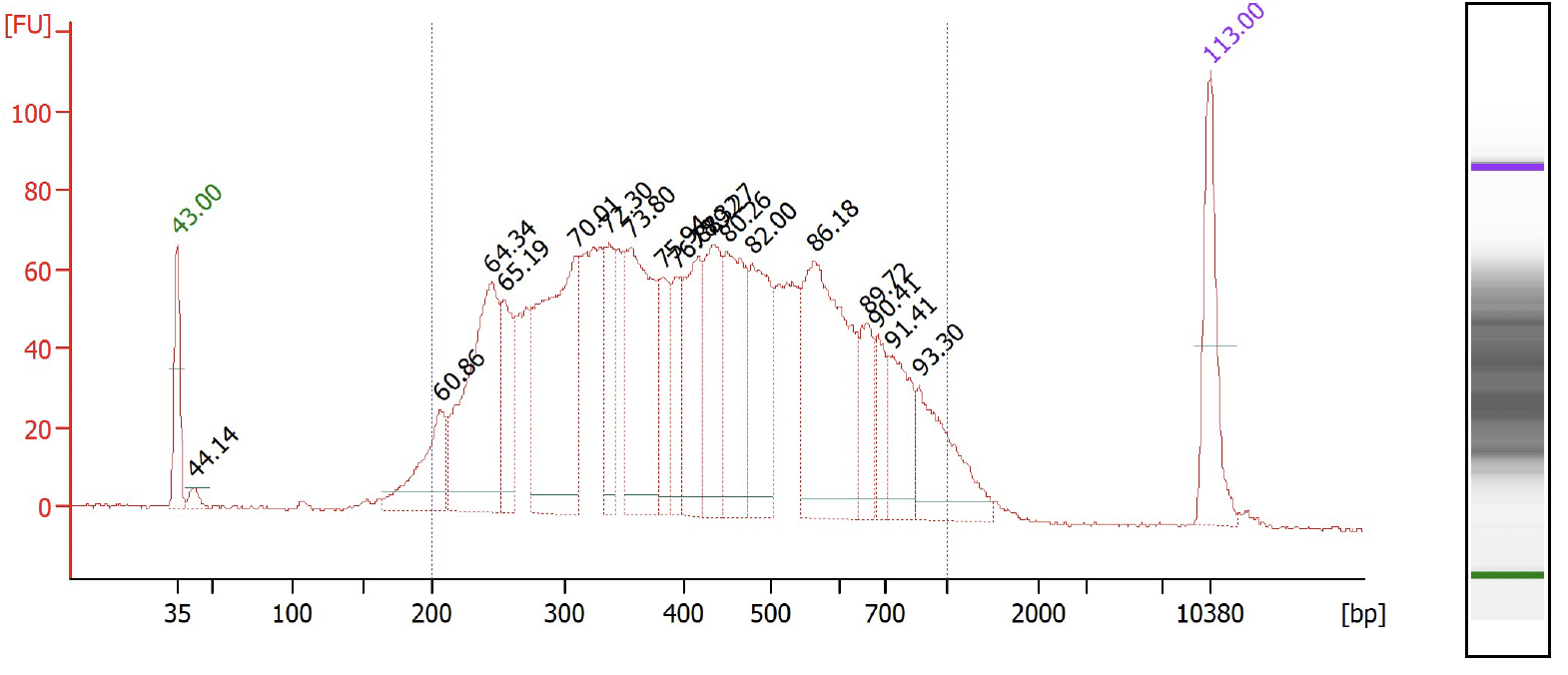
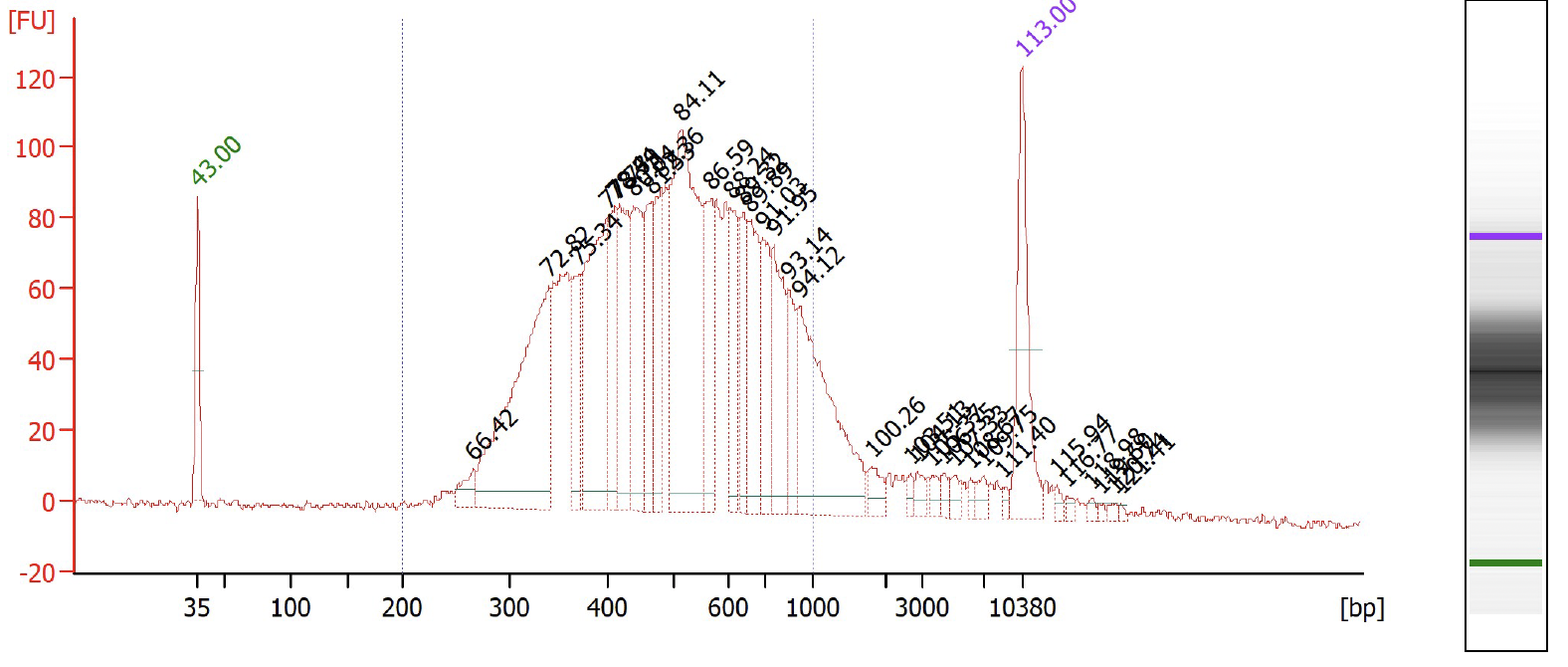
Check the RPE PCR cDNA library content and quality on a Agilent Bioanalzyer, using High Sensitivity DNA Analysis chips and concentration of the library using Qubit Fluorometric Quantification.
Indexed library preparation
Preparations:
Things to prepare:
- Thaw KAPA HiFi HotStart on ice
- Thaw the OAK-MO-primer-X.X primer
10micromolar (µM)(Refer Table S1) - Thaw the WTA1*R primer
10micromolar (µM)(Refer Table S1)
Estimate the molarity of the RPE PCR cDNA (from step 45 ):
-
Average fragment size (estimated with BioAnalyzer).
-
Library concentration (Qubit measurement)
Library concentration (nM): $$
Prepare 10nM normalized RPE cDNA library for index PCR
Prepare Index PCR mix:
Note: The KAPA HiFI DNA polymerase with hot start has very low 3-5' exonuclease activity. Still it is recommended to prepare the mix on ice.
| A | B | C | D | E |
|---|---|---|---|---|
| Stock | Unit | Final | Volume | |
| KAPA HiFi Hotstart ready mix | 2 | X | 1 | 25 |
| OAK-MO-primer-X.X | 10 | uM | 1 | 5 |
| WTA1*R | 10 | uM | 1 | 5 |
| Purified RPE product | 10 | nM | 2 | 10 |
| dH2O | 5 | |||
| 50 |
Prepare the PCR mix above and transfer 40µL of PCR master mix to the PCR tube having the normalized 10nanomolar (nM) RPE product.
Quick centrifugation to collect reaction to the bottom of the strip tube, pipette mix the reaction before running the following PCR program in a thermocycler for Index PCR amplification.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial Denaturation | 95*C | 3 minutes | 1x |
| Denaturation | 95*C | 30 seconds | |
| Annealing | 60*C | 30 seconds | 10 cycles |
| Elongation | 72*C | 30 seconds | |
| Final Elongation | 72*C | 5 minutes | 1x |
| Hold | 12*C | Hold | 1X |
Perform Ampure XP purification as below:
Important: Ensure to vortex and mix the Ampure XP beads well before adding for purification.
-
To purify cDNA add 0.8:1 ratio of Ampure XP beads to sample (
80µLof Ampure XP), and and mix by gently pipetting up and down. -
Incubate at
Room temperaturefor0h 5m 0s. -
Spin down and place on magnet and allow beads to be capture on the magnet. Place on magnet to separate the beads for
0h 3m 0s -
Discard supernatant, and wash once with
200µLof of freshly prepared 80% ethanol . After0h 0m 30sremove and discard the 80% ethanol. -
Repeat step 4 , 1X additional time.
-
Spin down the tube to bring down the beads to the bottom of the tube. Place on the magnet to remove the residual 80% ethanol.
-
Allow the bead pellet to dry for
0h 2m 0s. -
Elute cDNA in
100.5µLof Elution Buffer. Elute out100µLof final index PCR library to a fresh lo-bind300µLpcr strip tubes.
Perform another round of Ampure XP purification as below:
Important: Ensure to vortex and mix the Ampure XP beads well before adding for purification.
-
To purify cDNA add 1:1 ratio of Ampure XP beads to sample (
100µLof Ampure XP), and and mix by gently pipetting up and down. -
Incubate at
Room temperaturefor0h 5m 0s. -
Spin down and place on magnet and allow beads to be capture on the magnet. Place on magnet to separate the beads for
0h 3m 0s -
Discard supernatant, and wash once with
200µLof of freshly prepared 80% ethanol . After0h 0m 30sremove and discard the 80% ethanol. -
Repeat step 4 , 1X additional time.
-
Spin down the tube to bring down the beads to the bottom of the tube. Place on the magnet to remove the residual 80% ethanol.
-
Allow the bead pellet to dry for
0h 2m 0s. -
Elute cDNA in
60.5µLof Elution Buffer. Elute out60µLof final Index PCR product to a fresh lo-bind 1.5 ml eppendorf tube.
Check the sample index library content and quality on a Agilent Bioanalzyer, using High Sensitivity DNA Analysis chips and concentration of the library using Qubit Fluorometric Quantification.
Nova-ST library sequencing
The sequencing ready library can be sequenced on any Illumina compatible sequencers. The sequencing is performed as paired end sequencing. Sequencing parameter used for sequencing of the Nova-ST library is:
| A | B |
|---|---|
| Read Configuration | Number of bases |
| Read 1 | 33 |
| Index 1 | 8 |
| Index 2 | 8 |
| Read 2 | 80 |
Data processing
Details on data processing related to Nova-ST spatial processing can be found here: https://github.com/aertslab/Nova-ST