NEBNext® Varskip Short ARTIC SARS-CoV-2 RT-PCR Module E7626
Isabel Gautreau
Abstract
This protocol details methods for the NEBNext® ARTIC SARS-CoV-2 RT-PCR Module, NEB #E7626S/L 24/96 reactions.
cDNA Synthesis and Targeted cDNA Amplification with NEBNext VarSkip Short Primer Mixes:
This protocol follows an alternate variant-tolerant approach for targeting SARS-CoV-2 by utilizing NEBNext VarSkip Short SARS-CoV-2 Primer Mixes. The NEBNext VarSkip Short SARS-CoV-2 Primer mixes cannot be added to the same cDNA amplification reaction as the NEBNext ARTIC SARS-CoV-2 Primer Mixes. If downstream applications include sequencing, performing RNA input normalization prior to cDNA synthesis and targeted amplification promotes more even distribution of reads across sequencing libraries.
For other NEBNext® ARTIC SARS-CoV-2 protocols, please see the NEBNext ARTIC Protocols Collection.
To obtain instructions for using NEBNext ARTIC SARS-CoV-2 Primer Mix and the NEBNext® ARTIC SARS-CoV-2 RT-PCR Module please see the NEBNext ARTIC SARS CoV2 RT PCR Module Manual.
Before start
The amount of RNA required for detection depends on the abundance of the RNA of interest. In general, we recommend, using > 10 copies of the SARS-CoV-2 viral genome as input. In addition, we recommend setting up a no template control reaction and that reactions are set-up in a hood.
The presence of carry-over products can interfere with sequencing accuracy, particularly for low copy targets. Therefore, it is important to carry out the appropriate no template control (NTC) reactions to demonstrate that positive reactions are meaningful.
Steps
cDNA Synthesis
Gently mix and spin down the LunaScript RT SuperMix reagent. Prepare the cDNA synthesis reaction as described below:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| RNA Sample | 8 µl |
| (lilac) LunaScript RT SuperMix | 2 µl |
| Total Volume | 10 µl |
For no template controls, mix the following components:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| (white) Nuclease-free Water | 8 µl |
| (lilac) LunaScript RT SuperMix | 2 µl |
| Total Volume | 10 µl |
Incubate reactions in a thermocycler* with the following steps:
| A | B | C | D |
|---|---|---|---|
| CYCLE STEP | TEMP | TIME | CYCLES |
| Primer Annealing | 25°C | 2 minutes | 1 |
| cDNA Synthesis | 55°C | 20 minutes | |
| Heat Inactivation | 95°C | 1 minute | |
| Hold | 4°C | ∞ |
*Set heated lid to 105°C
cDNA Amplification
Gently mix and spin down reagents. Prepare the split pool cDNA amplification reactions as described below:
For Pool Set A:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| cDNA (Previous Step) | 4.5 µl |
| (lilac) Q5 Hot Start High-Fidelity 2X Master Mix | 6.25 µl |
| NEBNext VarSkip Short SARS-CoV-2 Primer Mix 1 | 1.75 µl |
| Total Volume | 12.5 µl |
For Pool Set B:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| cDNA (Step 2) | 4.5 µl |
| (lilac) Q5 Hot Start High-Fidelity 2X Master Mix | 6.25 µl |
| NEBNext VarSkip Short SARS-CoV-2 Primer Mix 2 | 1.75 µl |
| Total Volume | 12.5 µl |
Incubate reactions in a thermocycler* with the following steps:
| A | B | C | D |
|---|---|---|---|
| CYCLE STEP | TEMP | TIME | CYCLES |
| Initial Denaturation | 98°C | 30 seconds | 1 |
| Denature | 95°C | 15 seconds | 35 |
| Annealing/Extension | 63°C | 5 minutes | |
| Hold | 4°C | ∞ | 1 |
*Set heated lid to 105°C
Combine the Pool A and Pool B PCR reactions for each sample.
Cleanup of cDNA Amplicons
Vortex NEBNext Sample Purification Beads to resuspend.
Add 20µL to the PCR reaction. Mix well by pipetting up and down at least 10 times. Be careful to expel all of the liquid out of the tip during the last mix. Vortexing for 3-5 seconds on high can also be used. If centrifuging samples after mixing, be sure to stop the centrifugation before the beads start to settle out.
Incubate samples on bench top for at least 0h 5m 0s at Room temperature.
Place the tube/plate on an appropriate magnetic stand to separate the beads from the supernatant.
After 5 minutes (or when the solution is clear), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Add 200µL to the tube/plate while in the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Repeat the previous step once for a total of two washes:
Add 200µL to the tube/plate while in the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Be sure to remove all visible liquid after the second wash. If necessary, briefly spin the tube/plate, place back on the magnet and remove traces of ethanol with a p10 pipette tip.
Air dry the beads for up to 5 minutes while the tube/plate is on the magnetic stand with the lid open.

Remove the tube/plate from the magnetic stand. Elute the DNA target from the beads by adding 16µL. Elution volume can be adjusted for specific applications.
Mix well by pipetting up and down 10 times, or on a vortex mixer. Incubate for at least 0h 2m 0s at Room temperature. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand.
Place the tube/plate on the magnetic stand. After 5 minutes (or when the solution is clear), transfer 14µL to a new PCR tube. If elution volume was adjusted, the transfer volume should also be adjusted.
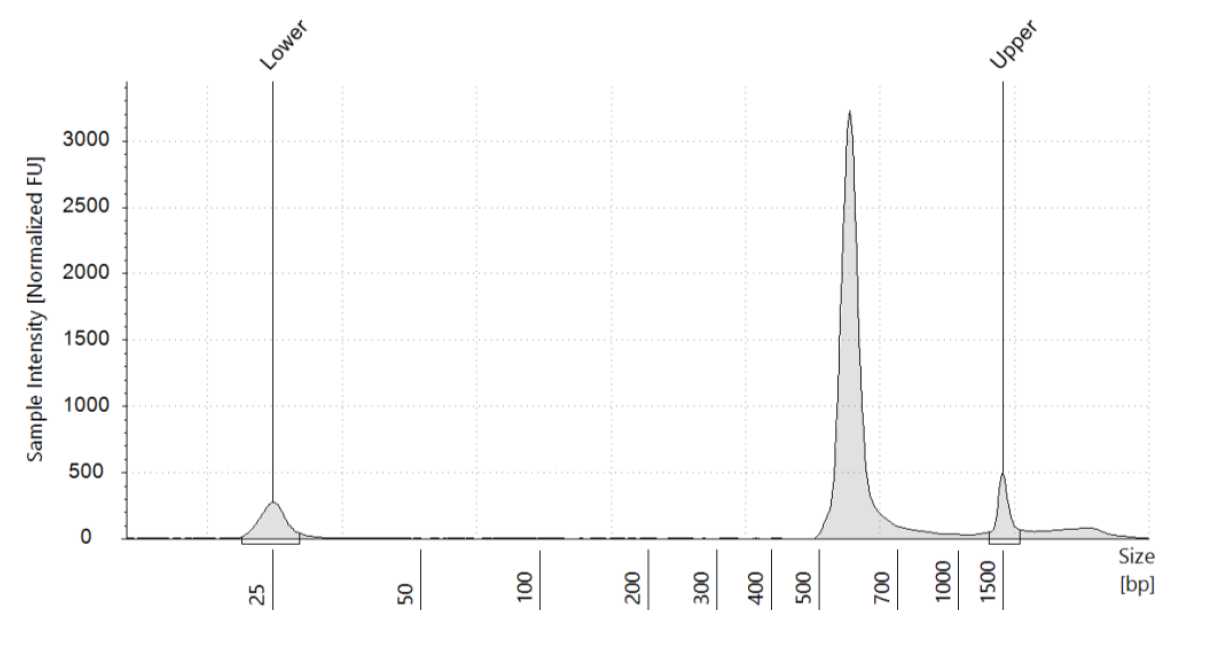
We recommend assessing the cDNA amplicon concentrations with a size distribution with Qubit® fluorometer.
Samples can be stored at –20°C for up to a week.