NCBI submission protocol for microbial pathogen surveillance
Ruth Timme, Julie Haendiges, Errol Strain, Maria Balkey, Tina Lusk Pfefer
Disclaimer
Please note that this protocol is public domain, which supersedes the CC-BY license default used by protocols.io.
Abstract
PURPOSE: Step-by-step instructions for submitting pathogen whole genome sequence data to NCBI and to the NCBI Pathogen Detection portal. This protocol covers the steps needed to establish a new NCBI submission environment for your laboratory, including the creation of new BioProject(s) and submission groups. Once these are step up, the protocol then walks through the process for submitting raw reads to SRA and sample metadata to BioSample through the Submission portal.
SCOPE: for use by any laboratory submitting WGS data for bacterial pathogens, including pathogens under surveillance at NCBI Pathogen Detection. (This includes US laboratories in GenomeTrakr, NARMS, Vet-LIRN, PulseNet, and other non-US networks and submitters).
For new submitters, there's quite a bit of groundwork that needs to be established before a laboratory can start its first data submission. We recommend that one person in the laboratory take a few days to get everything set up in advance of when you expect to do your first data submission.
If you need a pipeline for frequent or large volume submissions, follow Step 1 to get your NCBI submission environment established, then contact gb-admin@ncbi.nlm.nih.gov to set up an account for submitting through the API.
This protocol covers submission using NCBI's Submission Portal web-interface.
Version history:
V4: updated screenshots to reflect NCBI submission portal changes. Updated custom BioSample template.
V5 : Linking directly to the metadata template guidance instead of including duplicate copies of the files in this protocol. Updated screenshot for choosing the pathogen template to reflect changes at NCBI.
V6 : minor edits including updating links out to other protocols.
V7: Updated guidance for creating new BioProjects, including projects for non-targeted species
V8: Corrected the multi-species umbrella project accession in Step 1.5
Before start
This protocol has three sections:
- Section 1: Setting up NCBI accounts (for new users)
- Section 2: Single-step data submission to SRA for raw reads and associated sequence metadata and to BioSample for sample metadata
- Section 3 : Detailed steps for creating a BioProject (usually done once during the account set-up)
Associated protocols:
- Guidance for populating GenomeTrakr metadata templates (BioSample and SRA)
- NCBI Data Curation protocol for making updates, corrections, or retractions to your data.
Steps
Establish submission environmnet at NCBI
Set up a new NCBI submission environment for your lab:
1.1: Create an NCBI user account
1.2: Set up an NCBI submission user group for your lab
1.4 : Bookmark the link to your submission portal
1.5 . Identify or establish new BioProjects (detailed in Step 3 )
Ready for data submission:
After these steps are complete you can proceed with data submission in Step 2 .
Create an NCBI user account at NCBI: https://www.ncbi.nlm.nih.gov/account. This will be your own individual user account at NCBI.
***NCBI login changes updated in June, 2021. Read more here.
Establish an NCBI submission user group for your laboratory.
We recommend using this user group for all NCBI submissions related to microbial genome surveillance. This will link your laboratory's NCBI data ownership to the user group and not to individuals, allowing anyone in the current group to perform updates or retractions and answer inquiries from the NCBI staff, even if there's been a complete turnover of staff since the original data submission.
User groups also ensure consistent data ownership across BioProjects, BioSamples, and sequence data. If your laboratory has non-overlapping research groups submitting and managing data at NCBI, multiple user groups can be established to track these efforts separately.
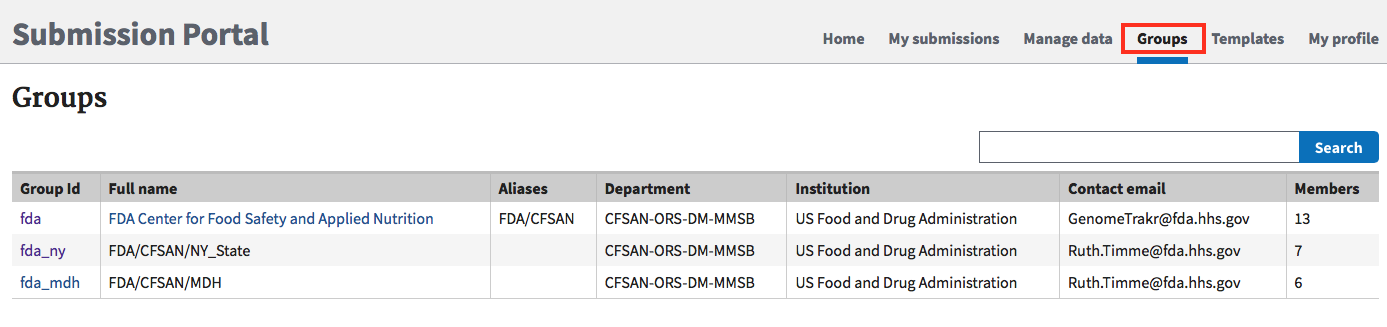
Your laboratory might already have a submission group established! Check the "Group" tab in the submission portal, https://submit.ncbi.nlm.nih.gov/groups/. Ask your colleagues to do the same thing, to ensure your laboratory doesn't already have one in place.
Creating a new submission group:
1 . Submit an email request to submit-help@ncbi.nlm.nih.gov containing the following information:
2 . Look for an email reply entitled "NCBI Submission Portal Group invitation" and click on the enclosed link to accept the invitation.
Managing your NCBI submission user group.
After a user group has been established it can be edited for membership and permissions by clicking in the “group” tab of the submission portal (https://submit.ncbi.nlm.nih.gov/groups/), then on the Group Id hyperlink, e.g 'fda_ny' in the above example.
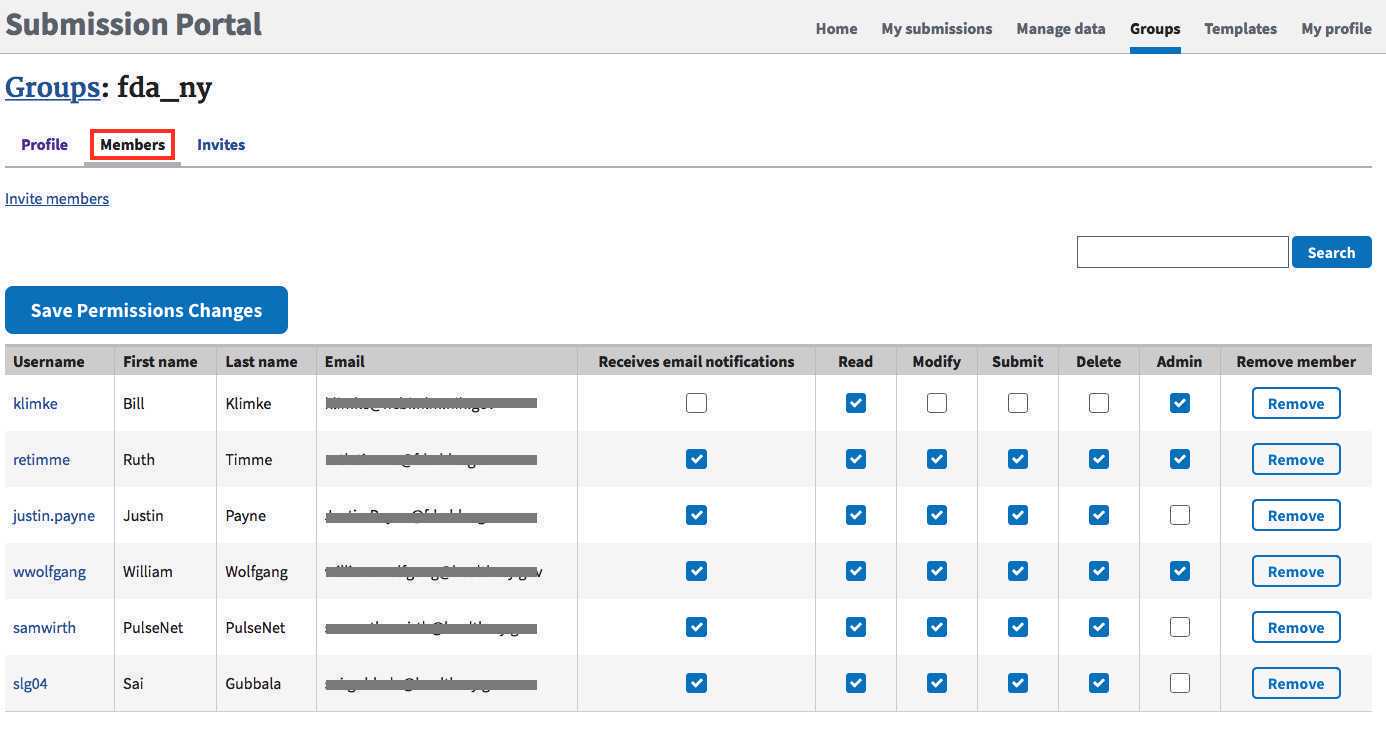
Users with admin privileges can update contact information in the "profile" tab and membership in the "Members" tab. New members can be invited by clicking on the "Invite members" link.
This user group should be kept up-to-date as members enter and leave the laboratory.
Permissions levels:
- READ: primarily for collaborators who can see the submissions, but not edit them.
- MODIFY, SUBMIT, DELETE: Permissions to submit, modify, or retract data (members usually have all or none of these permissions)
- ADMIN: Can invite or remove members of the submission group. Ensure that at least one (or more) members of your group have ADMIN privileges.
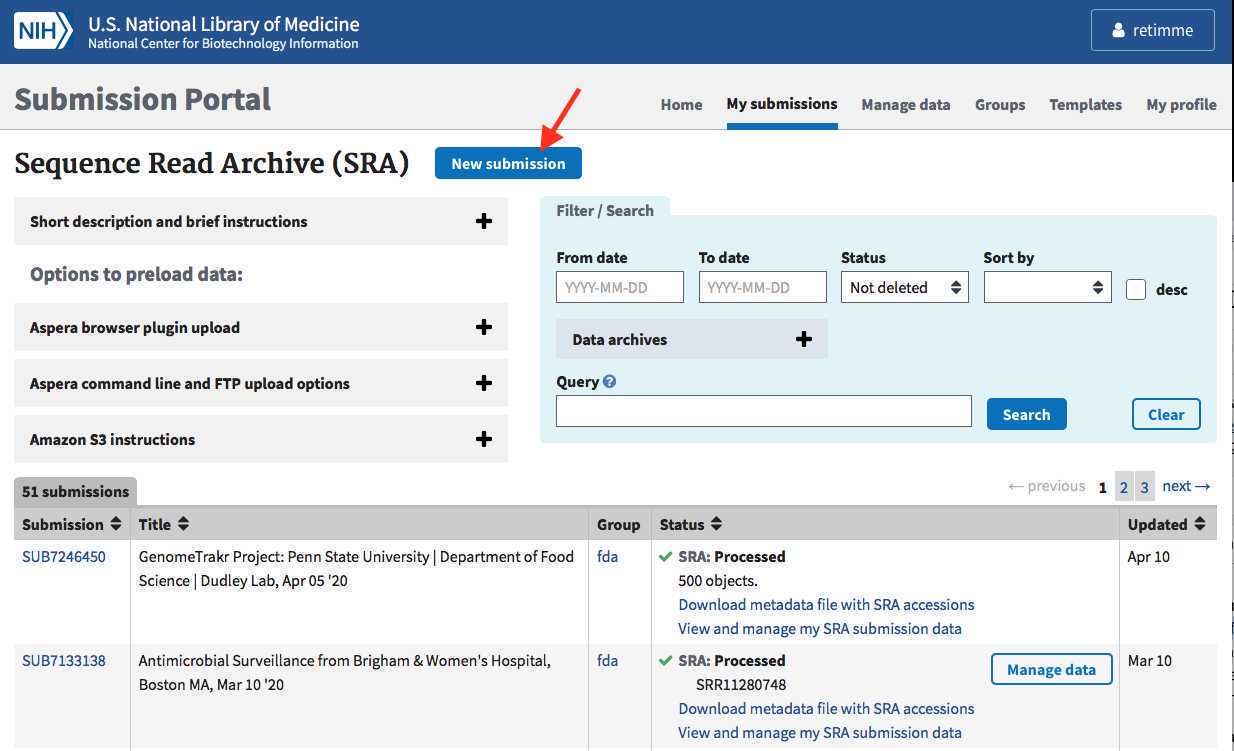
Bookmark “my submissions” at NCBI: https://submit.ncbi.nlm.nih.gov/subs/. This is the page where you view and track all of your past submissions.
If you see a blank page with a yellow box in the upper right corner saying “please login”, click this link and login using the credentials created in Step 1.1 .
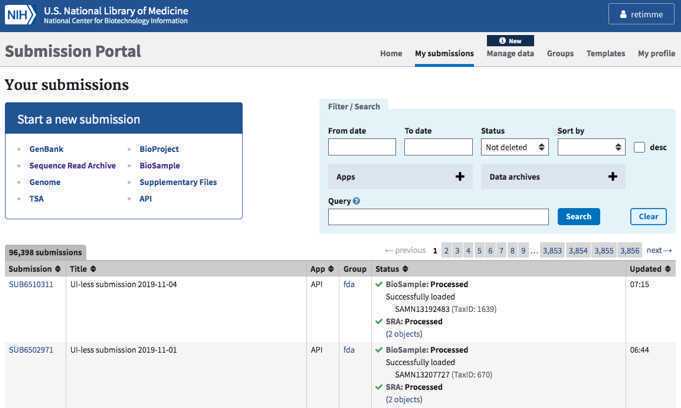
Identify or establish new BioProjects (Umbrella and/or Data BioProjects)
Umbrella BioProjects . If you are already part of a surveillance network, (e.g. GenomeTrakr, NARMS, Vet-LIRN, or PulseNet) you will create a new data project linked to an established umbrella BioProject. For reference, here is a list of the major Umbrella projects for GenomeTrakr and Vet-LIRN, organized by taxonomic classification. For species not included in this list, create a general non-targeted data BioProject for your lab linked to PRJNA593772 (our multi-species BioProject).
GenomeTrakr Umbrella projects ( https://www.ncbi.nlm.nih.gov/bioproject/PRJNA593772 ):
- Salmonella sp. PRJNA183844
- Listeria sp. PRJNA514048
- Escherichia coli / Shigella PRJNA230919
- Vibrio parahaemolyticus PRJNA245885
- Campylobacter sp. PRJNA258021
- Clostridium botulinum PRJNA290488
- Cronobacter sp. PRJNA258402
- All other species PRJNA706684
Vet-LIRN Umbrella projects:
- Salmonella enterica PRJNA314607
- Escherichia coli and Shigella PRJNA316449
- Staphylococcus PRJNA316451
If you need to establish a new umbrella BioProject, follow instructions in Step 3 with modifications for creating a new Umbrella BioProject ( Step 3.12 ).
Data BioProjects. Does your laboratory have an established data BioProject for this effort? If not please follow the instructions in Step 3 for creating a new one.
Data submission (BioSample and SRA)
Data submission (source metadata and sequence data):
This protocol follows a one-step data submission process where the source metadata is submitted through the same submission workflow as the sequence data.
**Before submission , ensure that your sequences meet the quality control (QC) thresholds for your surveillance network. You can follow your own internal QC process or use FDA's free GalaxyTrakr platform:
Quality control assessment for microbial genomes: GalaxyTrakr MicroRunQC workflow
Navigate to the My Submissions page in the NCBI Submission Portal: https://submit.ncbi.nlm.nih.gov/subs/
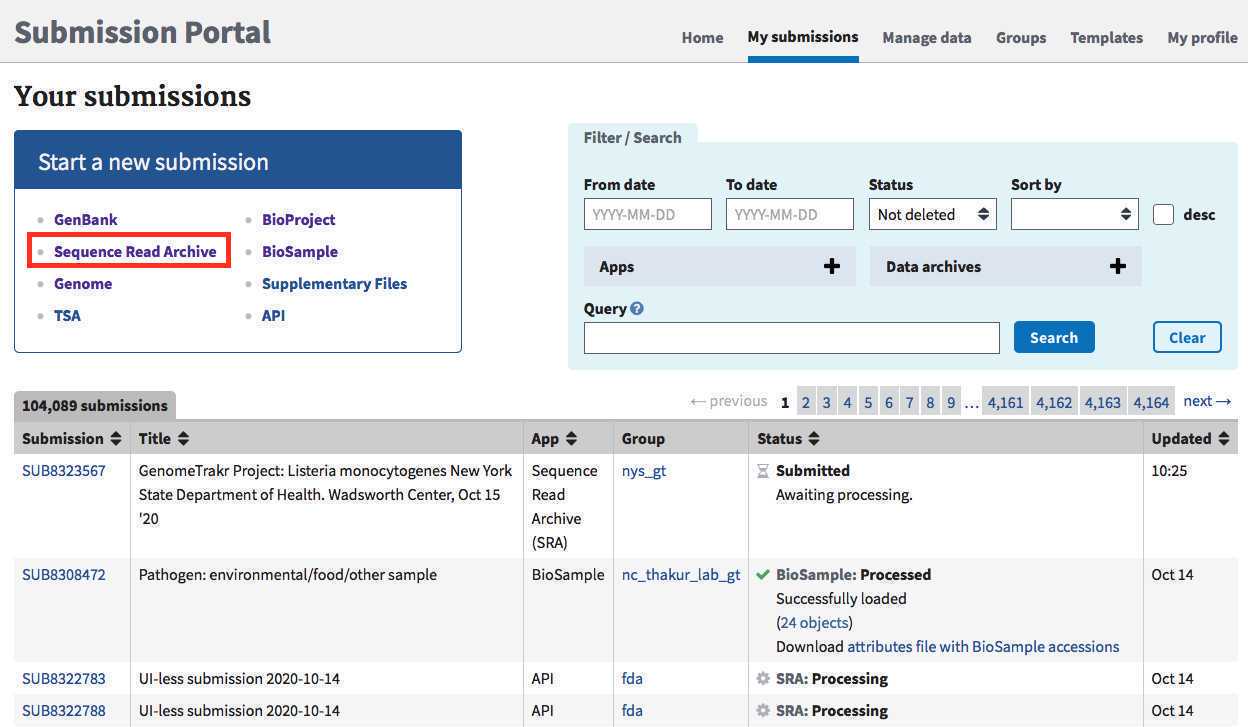
Click "Sequence Read Archive" to start a submission.
Download and populate the sample (BioSample) and sequence (SRA) metadata templates:
Custom metadata templates and guidance are available in the following protocol:
gui
Guidance for populating GenomeTrakr metadata templates (BioSample and SRA)
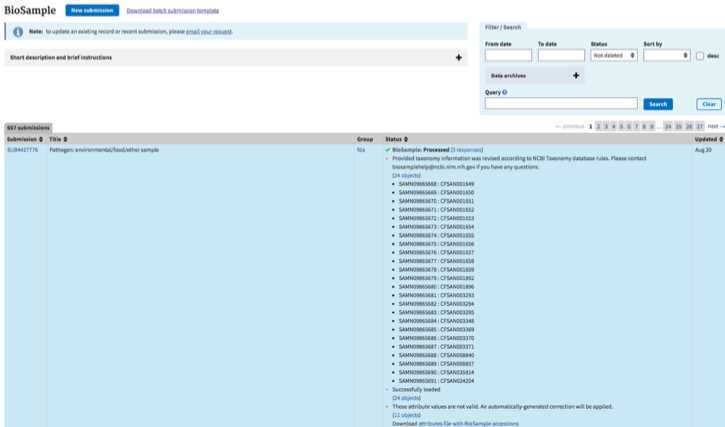
BioSample accessions:
BioSample accessions will be automatically created upon submission and will be available on the “my submissions” page of the Submission portal by clicking on “## objects” within the submission record. You can also download by clicking the “Download attributes file with BioSample accessions”. Accessions will start with SAMNxxxxxxxx. You will also receive an email within 12 hours, but typically much faster, containing these same accessions.
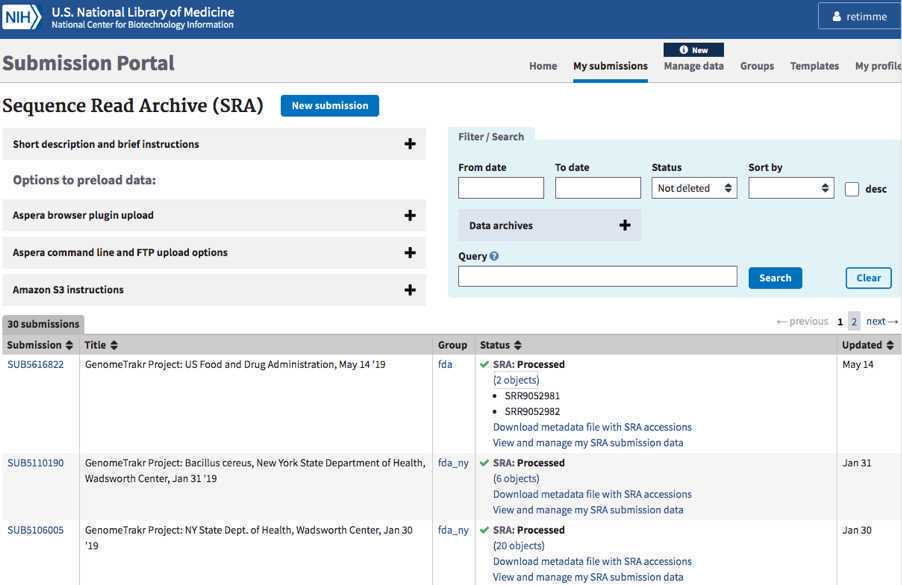
SRA Accessions:
SRA run accessions will be available on the “my submissions” page of the Submission portal by clicking on “## objects” within the submission record. You can also download by clicking the “Download metadata file with SRA accession”. Accessions will start with SRRxxxxxxx.” You will also receive an email with these same accessions within 24 hours, but typically much faster, containing these same accessions.
Important data stewardship and curation notes:
-
Develop an internal meIDthod for storing and tracking your BioSample and SRR accessions! They are required for making future updates to your records.
-
For updates, corrections, or retractions to your BioSample and SRA records, follow the guidance provided in the NCBI Curation Protocol. Some edits can be made within the submission portal and others need to be done via email.
NCBI data curation protocol - SOP for editing GenomeTrakr submissions
SUBMITTER tab:
Populate with submitter info. The “submitter” is the name of the person, or user group, who is physically doing the submissions, not a supervisor or PI.
Select the appropriate submission group name (see Step 1.2 for creating a new submission group), and describe the submitting organization or laboratory name. This will be auto-populated from the contact info you included in your NCBI user account. Click "Continue" to proceed.
GENERAL INFO tab:
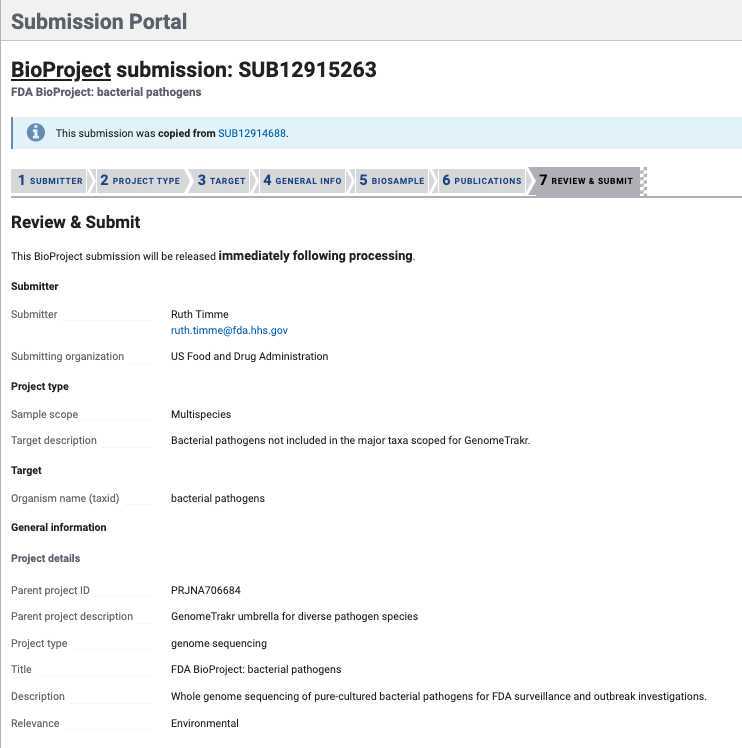
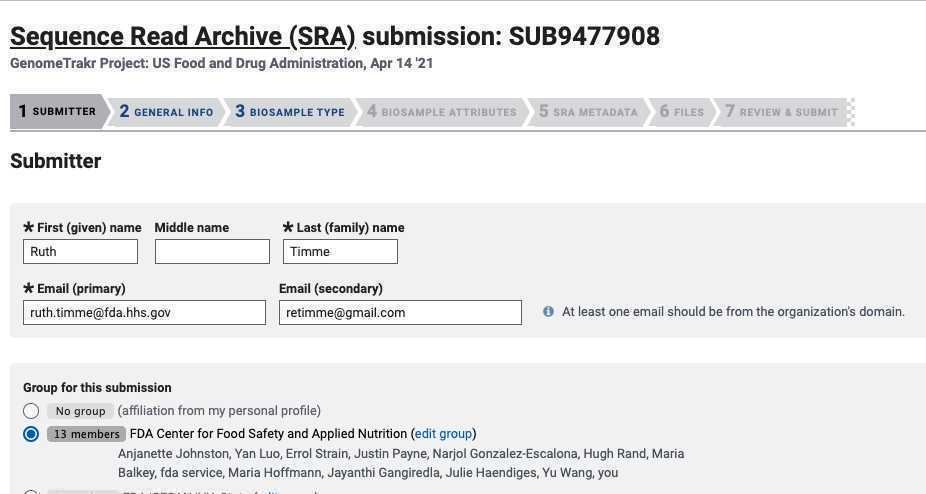
1. BioProject: Did you already have a data BioProject for this effort? If not please follow instructions in Step 3 for creating a new data or umbrella BioProject. Return back to this sub-step with the data BioProject accession in hand.
Click " Yes " and paste in your data BioProject accession, e.g. PRJNA614995.
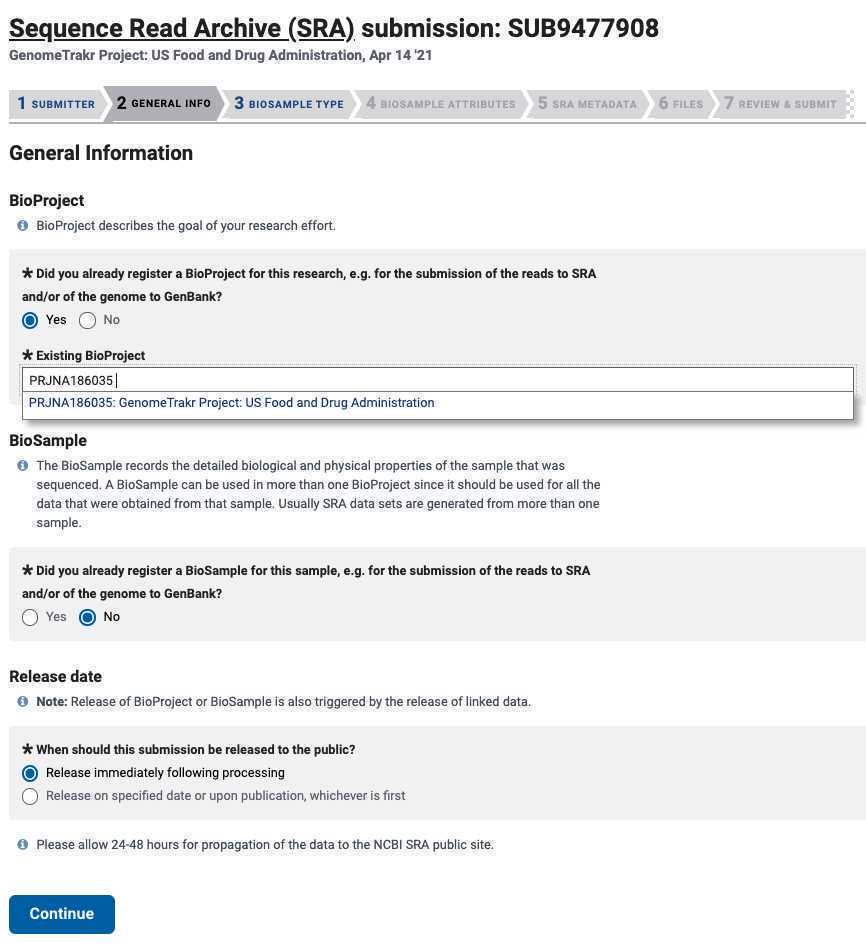
2. BioSample : Click "NO" here. You will be registering BioSamples within this current submission
3. Release date : Choose "Release immediately following processing".
- Click Continue .
BIOSAMPLE ATTRIBUTES tab:
Choose "Upload a file using Excel or text format (tab-delimited) that includes the attributes for each of your BioSamples".
Then click "Choose File" and browse to your populated metadata template.
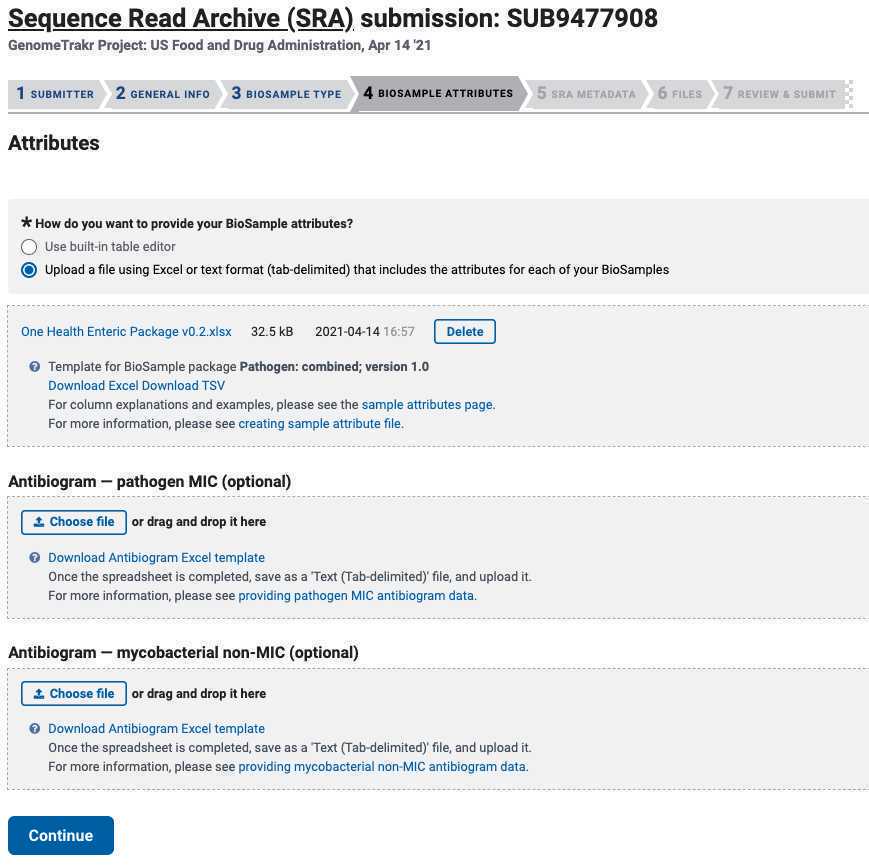
Antibiogram data : please provide if you have it!
Click " Continue ".
NCBI will do a validation check on your metadata. Resolve any red "errors" reported back by editing the spreadsheet and replacing the uploaded file. Review any yellow "Warnings" and proceed if everything looks ok.
Click " Continue ".
SRA METADATA tab:
Choose : "Upload a file using Excel or text format (tab-delimited)".
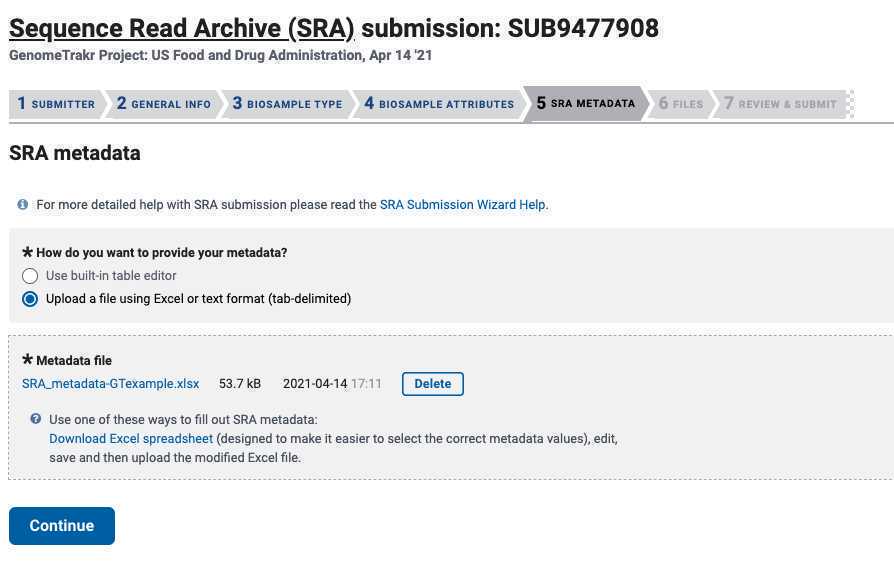
Upload your populated SRA metadata template (see Step 2.1 for where to get this file)
Click " Continue ".
NCBI will do a validation check on your sequence metadata. Resolve any red "errors" reported back by editing the spreadsheet and replacing the uploaded file. Review any yellow "Warnings" and proceed if everything looks ok.
Click " Continue ".
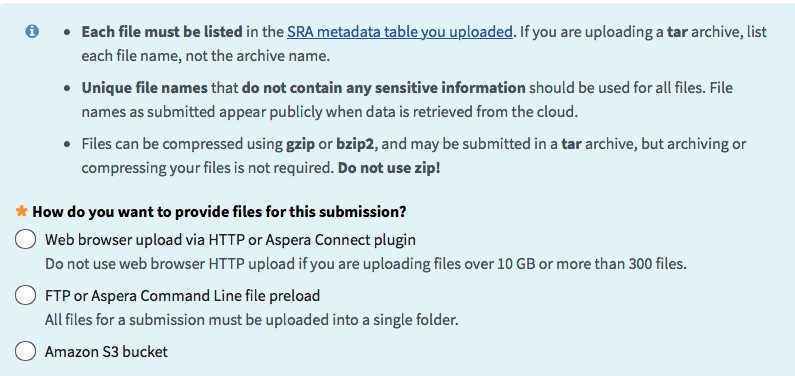
Files tab:
Each laboratory will establish its own path for transferring files.
In general, selecting the web browser option should work for uploading ~48 sequences at a time. For a more stable internet connection, your laboratory can use FTP or Aspera. Directions for doing so pop up after clicking the FTP radio button.
REVIEW & SUBMIT tab:
Check over your entire submission, then click submit.
If corrections are needed, you can go back and select individual tabs to edit your submission.
BioProject Creation
Create a new BioProject
BioProjects are an organizing tool at NCBI that pulls together different kinds of data submitted across multiple NCBI databases. Each BioProject has a unique URL, providing a home page with a title, description, links to lab websites, publications, funding resources associated with a particular project, along with links to the deposited data. A basic data BioProject holds actual sequence data, assemblies, and their associated metadata. An umbrella BioProject is a way to group two or more data BioProjects together, which is useful for coordinating disease surveillance and for looking across the grouped BioProjects in a single view.
This protocol describes the steps for creating a new data BioProject linked to an existing umbrella BioProject (usually established by a coordinating group, e.g. GenomeTrakr, NARMS, Vet-LIRN).
*If you need to create a new Umbrella BioProject, modifications are summarized in Step 3.12.
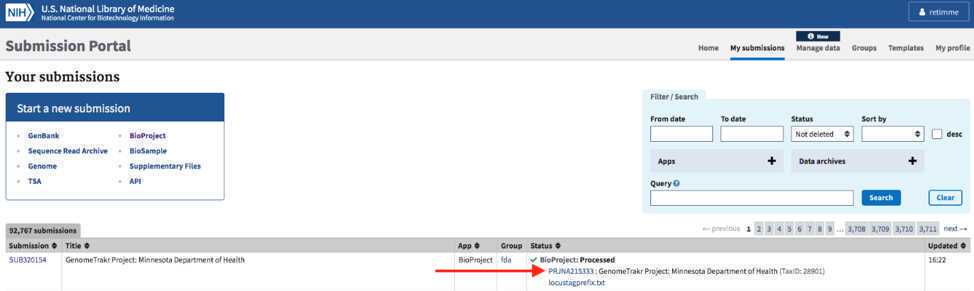
Navigate to the “My Submissions” page, https://submit.ncbi.nlm.nih.gov/subs/, and click “BioProject” in the “Start a new submission” box.
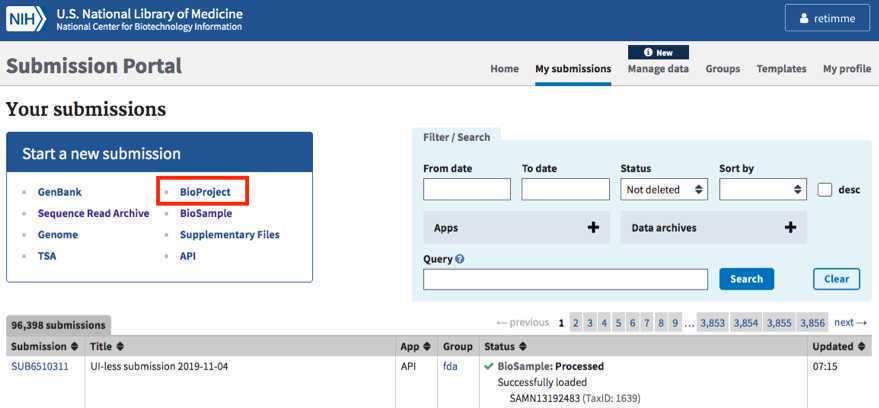
If you are part of a coordinated surveillance effort, like GenomeTrakr, please alert the coordinating body that a new BioProject was created under an existing umbrella.
For GenomeTrakr, contact genomeTrakr@fda.hhs.gov
Creating a new Umbrella BioProject:
Proceed as outlined in the above steps with the following modifications:
PROJECT TYPE tab:
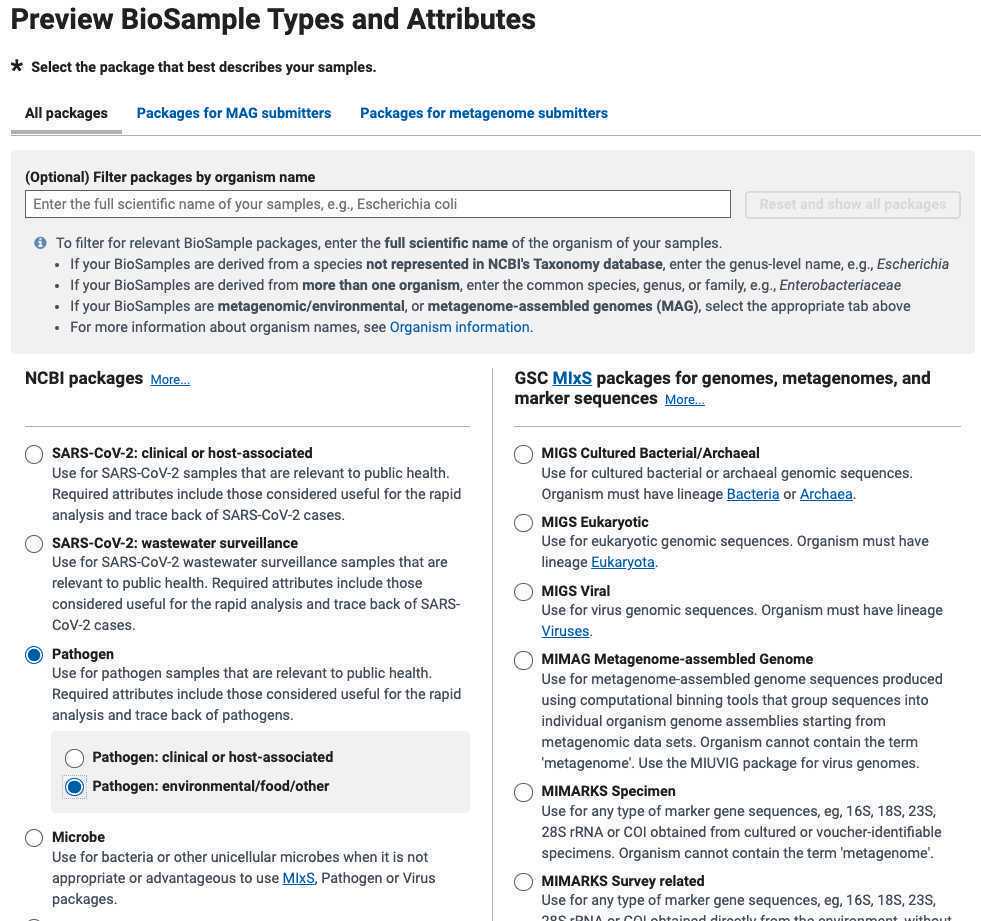
For an Umbrella BioProject : select multi-species. This will allow you to link multiple data BioProjects representing different species under a single umbrella.
TARGET tab:
For an Umbrella BioProject : Leave the Organism name field blank. Include a list or description of species you intend to include in this effort. E.g. “bacterial foodborne pathogens”, or “SARS-Cov-2”
GENERAL INFO tab:
Umbrella BioProject Title: e.g. "Microbial pathogen surveillance at NY State Dept. of Health, Wadsworth Center."
Is your project part of a larger initiative that is already registered at NCBI?
- For an Umbrella BioProject: click “NO”
The last step is to email _bioprojecthelp@ncbi.nlm.nih:_ bioprojecthelp@ncbi.nlm.nih:
Example email:
After the conversion is complete you can use the new Umbrella accession to properly link any new data BioProjects being created.
Important data stewardship and curation notes:
-
Develop an internal method for storing and tracking your BioProject accessions! They are required for every BioSample and sequence data submission to ensure proper linkage.
-
Bookmark URLs for each of your data BioProjects to monitor the public-facing view of your submissions. 5.g. Virginia DCLS's GenomeTrakr Salmonella BP: https://www.ncbi.nlm.nih.gov/bioproject/219491
-
For updates to your BioProjects, follow the guidance provided in the NCBI Curation Protocol. Some edits can be made within the submission portal and others need to be done via email. NCBI data curation protocol - SOP for editing GenomeTrakr submissions
Submitter tab:
Populate with submitter info. An NCBI "submitter” is the name of the person or submission group who is managing the submissions, not a supervisor or PI.
Select the appropriate submission group name (see Step 1.2 for creating a new submission group), and describe the submitting organization or laboratory name. This will be auto-populated from the contact info you included in your NCBI user account.
Project type tab:
Project data type: Genome sequencing and assembly.
Sample scope:
For a Data BioProject Data BioProject: select multi-species. This will allow you to submit multiple different species to the BioProject.
Target tab:
For a Data BioProject Data BioProject : Populate ONLY the Organism name here:
For targeted-pathogen BioProjects:
Organism name = Include a Genus name, e.g., Salmonella sp .
For non-targeted pathogens
Organism name = "bacteria"
Leave the strain info and Description fields blank.
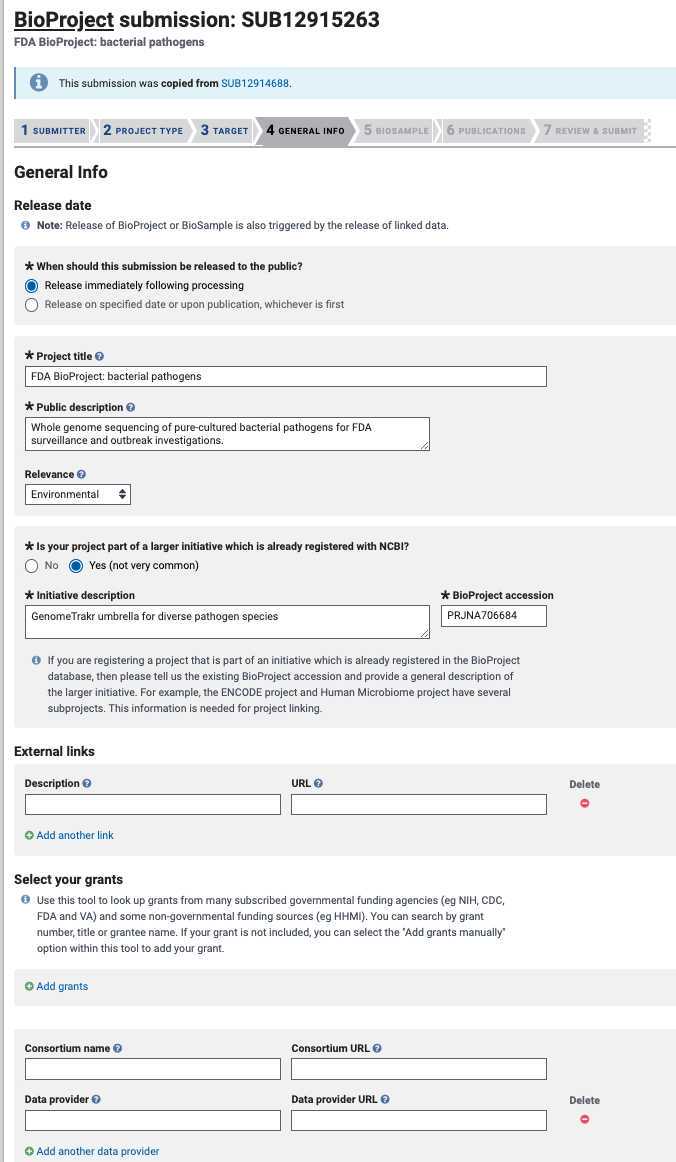
General info tab:
Click “Release immediately following processing”.
Include a brief title describing the effort.
- Data BioProject Data BioProject Title : e.g., “GenomeTrakr Project: NY State Dept. of Health, Wadsworth Center”.
Public Description: e.g., “Whole-genome sequencing of pure-cultured microbial pathogens as part of XXXX surveillance effort.”
Relevance: environmental.
Is your project part of a larger initiative that is already registered at NCBI?
- Data BioProjects . Click “Yes” and include a brief description and umbrella BioProject accession number (see Step 1.5 ). This will properly link your data project to the umbrella.
BioSample tab:
Leave blank!! You will create biosamples separately.
Publications tab:
If relevant, include publications from your laboratory.