NCBI data curation protocol - SOP for editing GenomeTrakr submissions
Ruth Timme, Errol Strain, Maria Balkey, Candace Hope Bias, Tina Lusk Pfefer
Disclaimer
This method is under development and assessment for suitability of use. It is likely that modifications will be made to improve the method.
Abstract
PURPOSE: After data are submitted to NCBI submitters often encounter the need to update, retract, or replace these records. This is called data curation. This protocol provides instructions for keeping these records up-to-date for each relevant database at NCBI.
SCOPE: This protocol covers curation for the following NCBI databases:
- BioProject
- BioSample
- Sequence Read Archive
Version history:
V2. Edit submissions using the NCBI portal (Manage data). Moved "how to find my data" content to a new protocol.
V3: Update to BioSample section, providing further guidance on updating taxonomic names.
V4: Clarifying protocol for SRA retraction
Before start
Most updates to existing NCBI submissions are performed through email requests to each respective NCBI database (e.g. BioSample, BioProject, Sequence Read Archive, and Pathogen Detection). NCBI curators within each respective database expect these emails to update and retract data. It is their job to help the data stay current, so do not hesitate to correct errors when they are spotted.
Steps
BioProject Curation
How to edit a BioProject
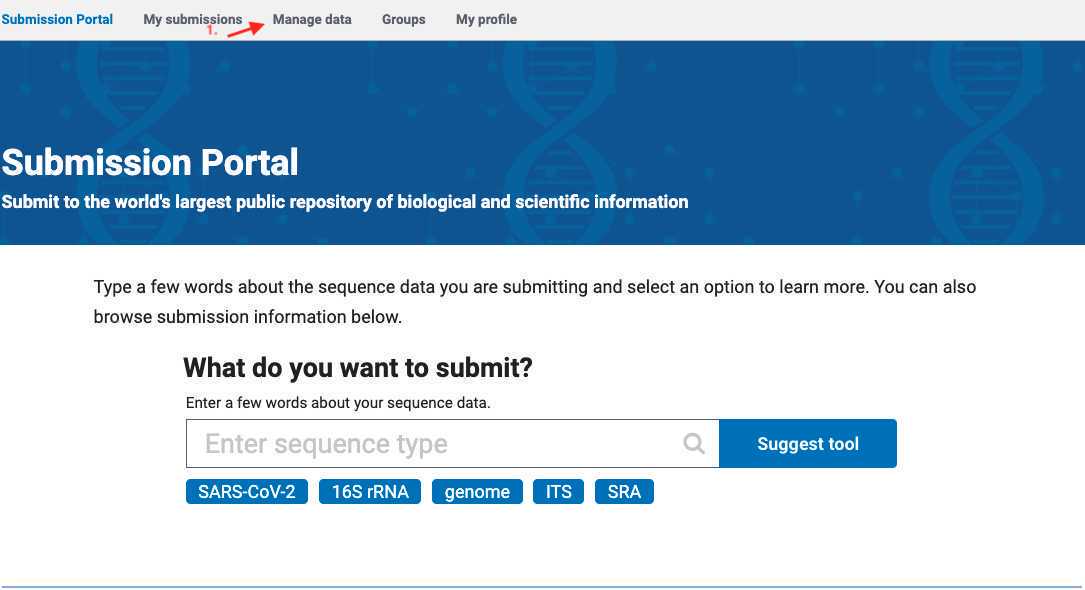
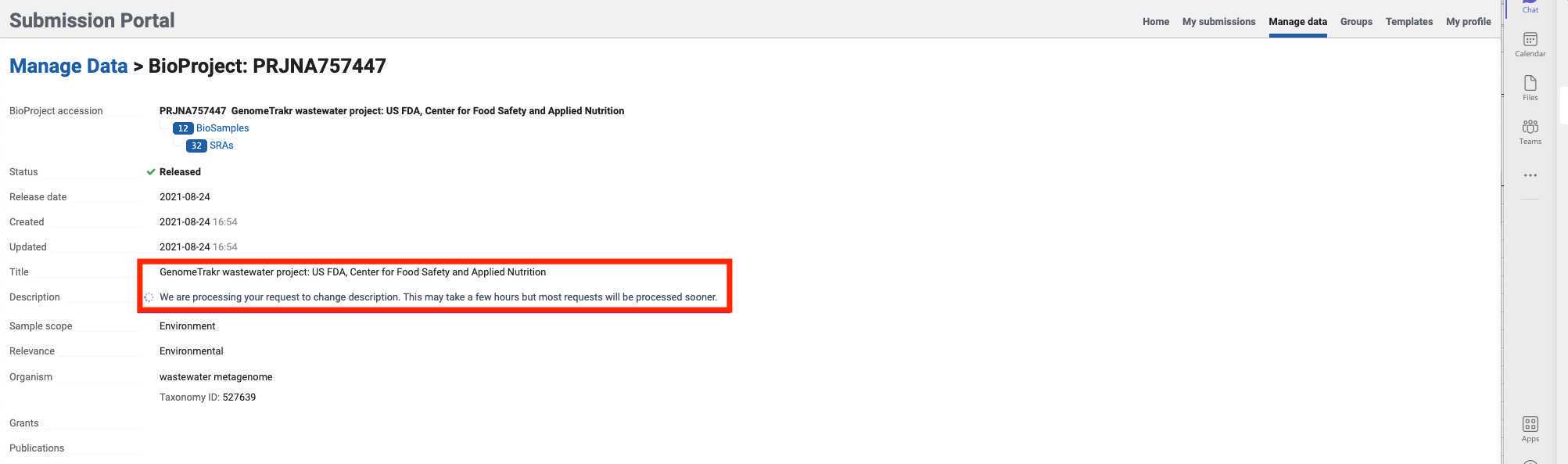
- Click on the "Manage Data" tab within the submission portal, or navigate directly to "Manage Data": https://dataview.ncbi.nlm.nih.gov to edit Title, Organism, Description, URL, or publications for your BioProject.
-
In the menu, select BioProject, a complete list of your NCBI group bioprojects will be displayed.
-
Click on the BioProject that you need to edit.
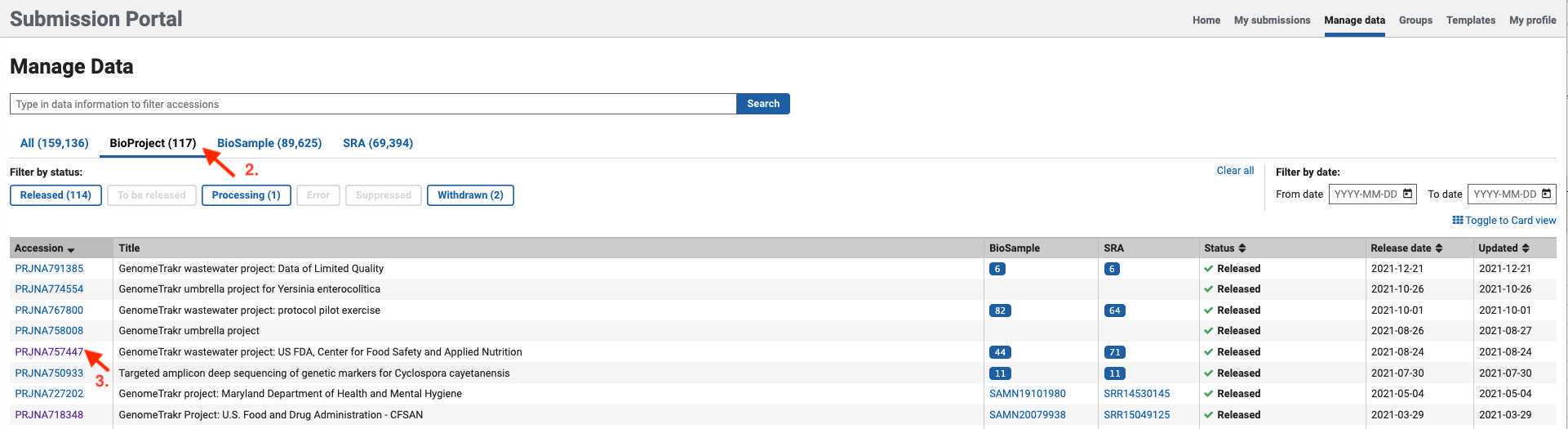
- Fields available for editing will be displayed after selecting a BioProject.

- Click in any of the edit/add fields and proceed to add the corresponding BioProject information. Once the information is changed or added, click next and submit.
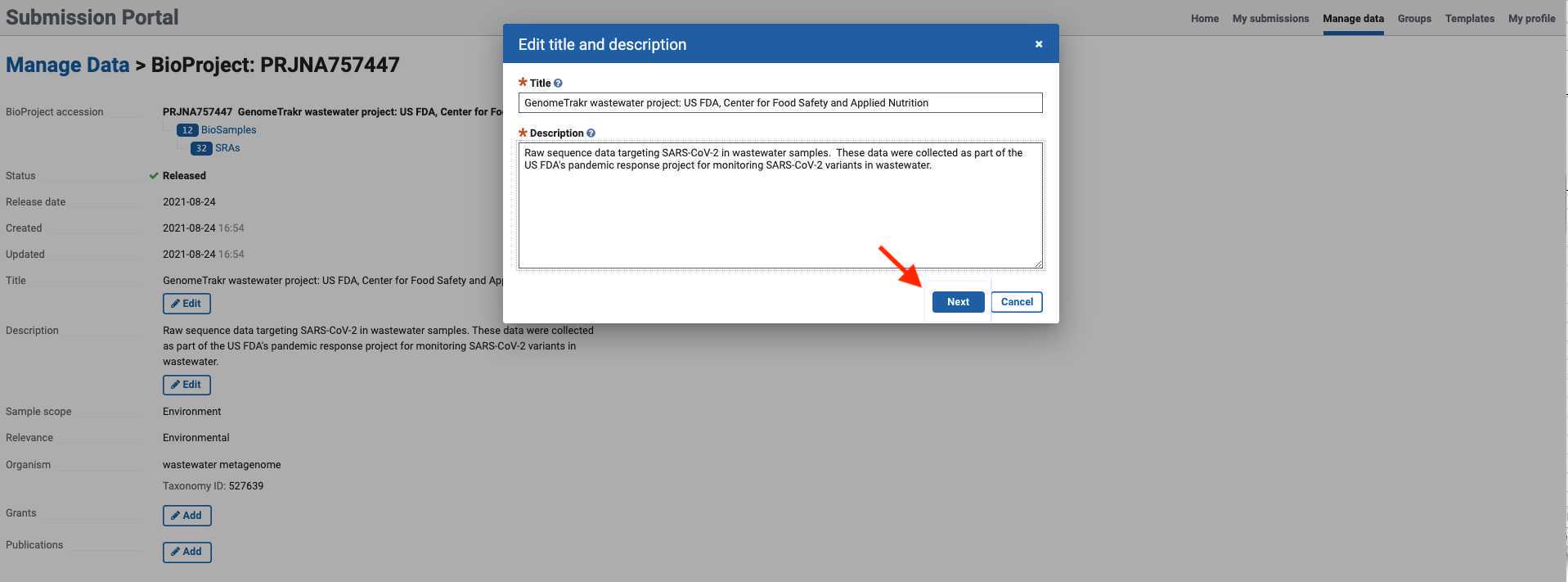
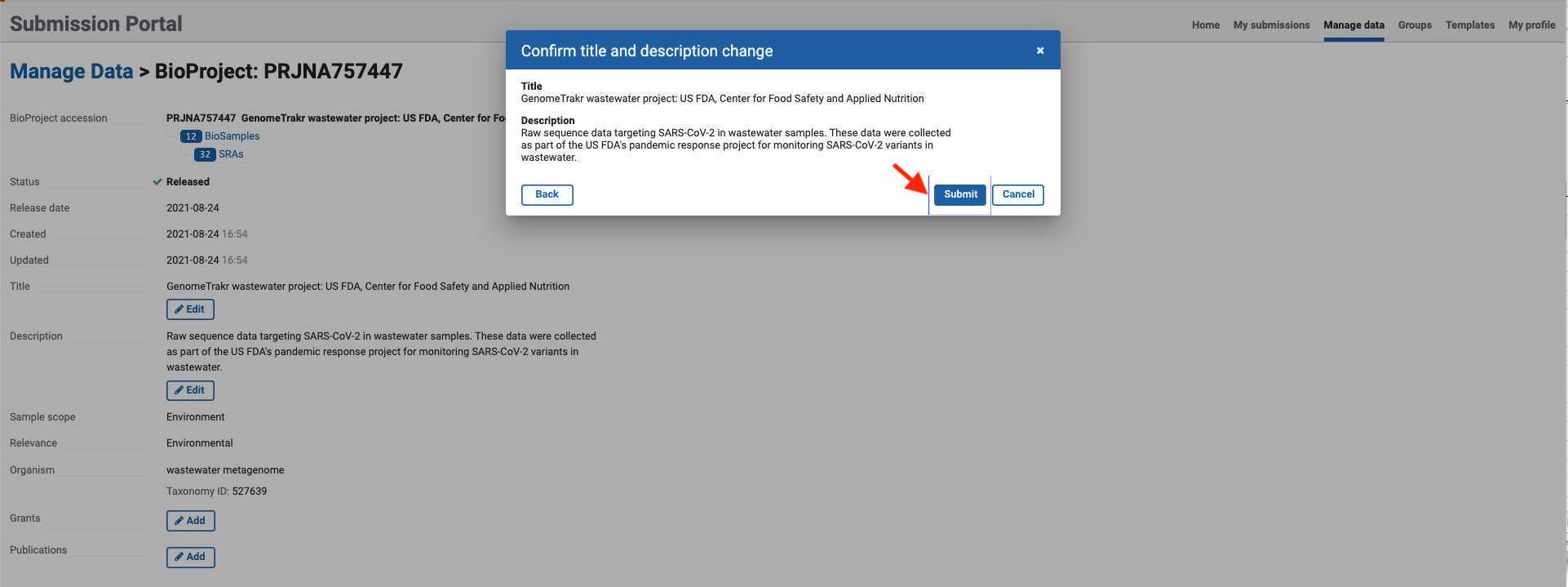
- A confirmation prompt will indicate that your updates are in progress.
Email for BioProject database: bioprojecthelp@ncbi.nlm.nih.gov bioprojecthelp@ncbi.nlm.nih.gov
Use this email for the following tasks, include the BioProject accession in the email subject:
-
Questions about errors or processing of a BioProject submission
-
Convert a Data BioProject to an Umbrella BioProject
-
Re-assign a BioProject from one Umbrella BioProject to another
BioSample curation
How to edit BioSamples.
All edits or updates to BioSample records are submitted via email to the BioSample database: biosamplehelp@ncbi.nlm.nih.gov. _biosamplehelp@ncbi.nlm.nih.gov._ biosamplehelp@ncbi.nlm.nih.gov.
Use this email for the following tasks. Include your lab and the request date in your subject line for easy tracking, eg “FDA BioSample update, Dec 10, 2019”.
-
Questions about validation errors or processing of a BioSample submission.
-
Update, correct, or add fields/attributes to a BioSample(s)
-
Retraction
-
Add a linkage or re-assign linkage to a BioProject
-
Add or change a strain or isolate field to an existing biosample where one has been lacking (necessary for the isolate's assembly to appear in GenBank).
-
Taxonomic updates: Include "pd-help@ncbi.hlm.nih.gov" on these requests to ensure taxonomic changes get propagated fully across NCBI databases. The organism name should include the Genus species, subspecies where present, plus serovar/serotype information. In cases where the BioSample attributes serovar/serotype were populated (e.g. with traditional serotyping results), ensure they are also updated as needed. Special note about Salmonella enterica isolates: please submit or update serotyping information in the serovar field, not the serotype field.
You will receive a confirmation email that the updates were performed. These types of transactions are common for this database, so do not hesitate to submit requests as needed.
How to retract one or multiple BioSamples
How to update content in metadata fields or add new fields/attributes to a BioSample record(s)
Attach a tab-delimited text file with the BioSample accessions in the first column and fields to update the right. You can attach a table to update one or multiple records at a time.
Examples:
FDA_biosample_update_20220203_fb.txt
(adding "sequenced_by" and "project_name" to a biosample)
- The following table will update the collection date and isolation source on one BioSample record: | A | B | C | | --- | --- | --- | | BioSample | collection_date | isolation_source | | SAMN12987335 | 2019-10-12 | cilantro |
Tab-delimited table for updating a BioSample record.
Re-assign a BioSample from one BioProject to another
Submit an update request with the new BioProject accession(s) specified in a column.
SRA curation
SRA updates and retractions
Make updates within the submission portal:
The following types of updates can be made within the submission portal under the “Manage data” tab:
- Sequence metadata, such as library ID, library strategy, sequencing platform or instrument.
- Associated BioSample or BioProject accession numbers
- Release date
-
Click on the "Manage Data" tab within the submission portal, or navigate directly to "Manage Data": https://dataview.ncbi.nlm.nih.gov
-
Query for SRR accession you'd like to update:
-
Click on the resulting "BioProject" link.
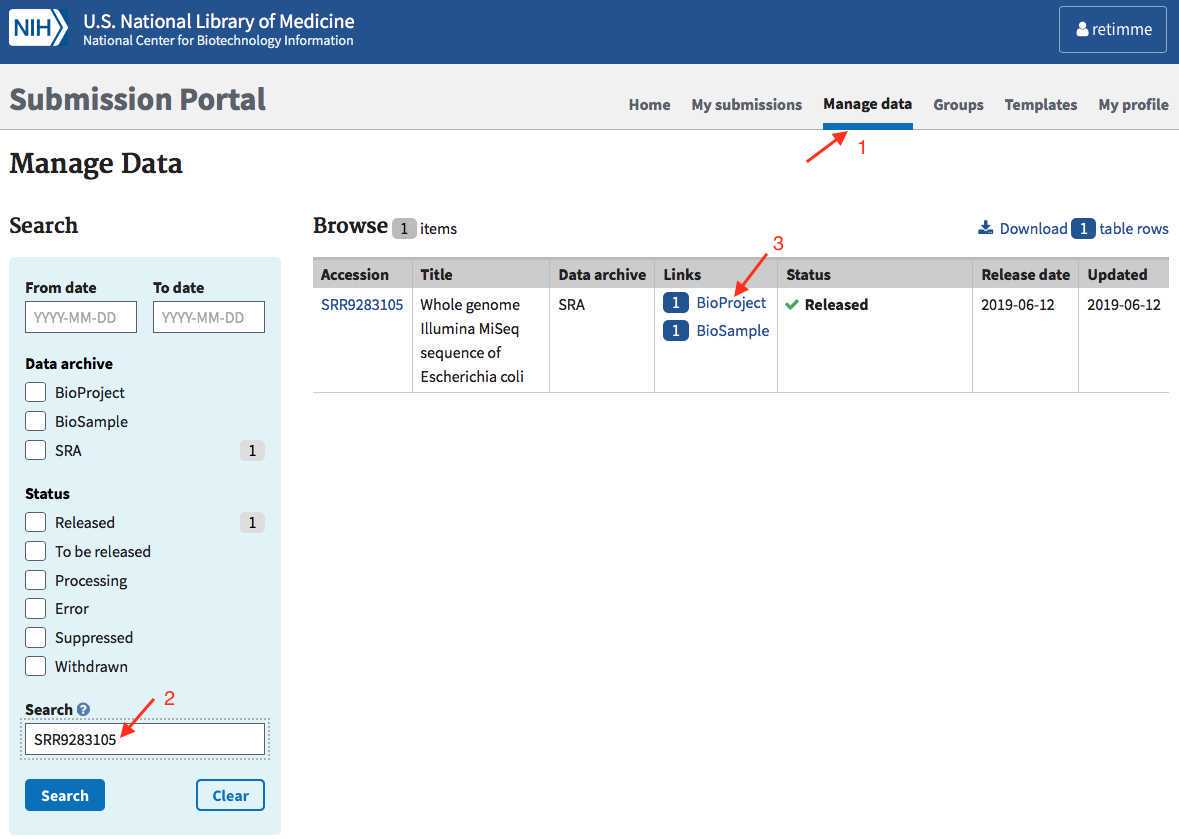
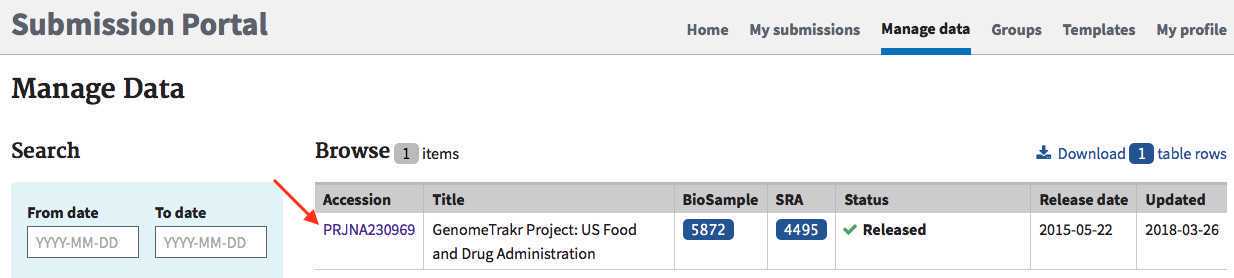
- Click on the BioProject accession link:
- All the SRA records submitted to this BioProject can now be edited! Search for the one(s) you want and click the box to edit.
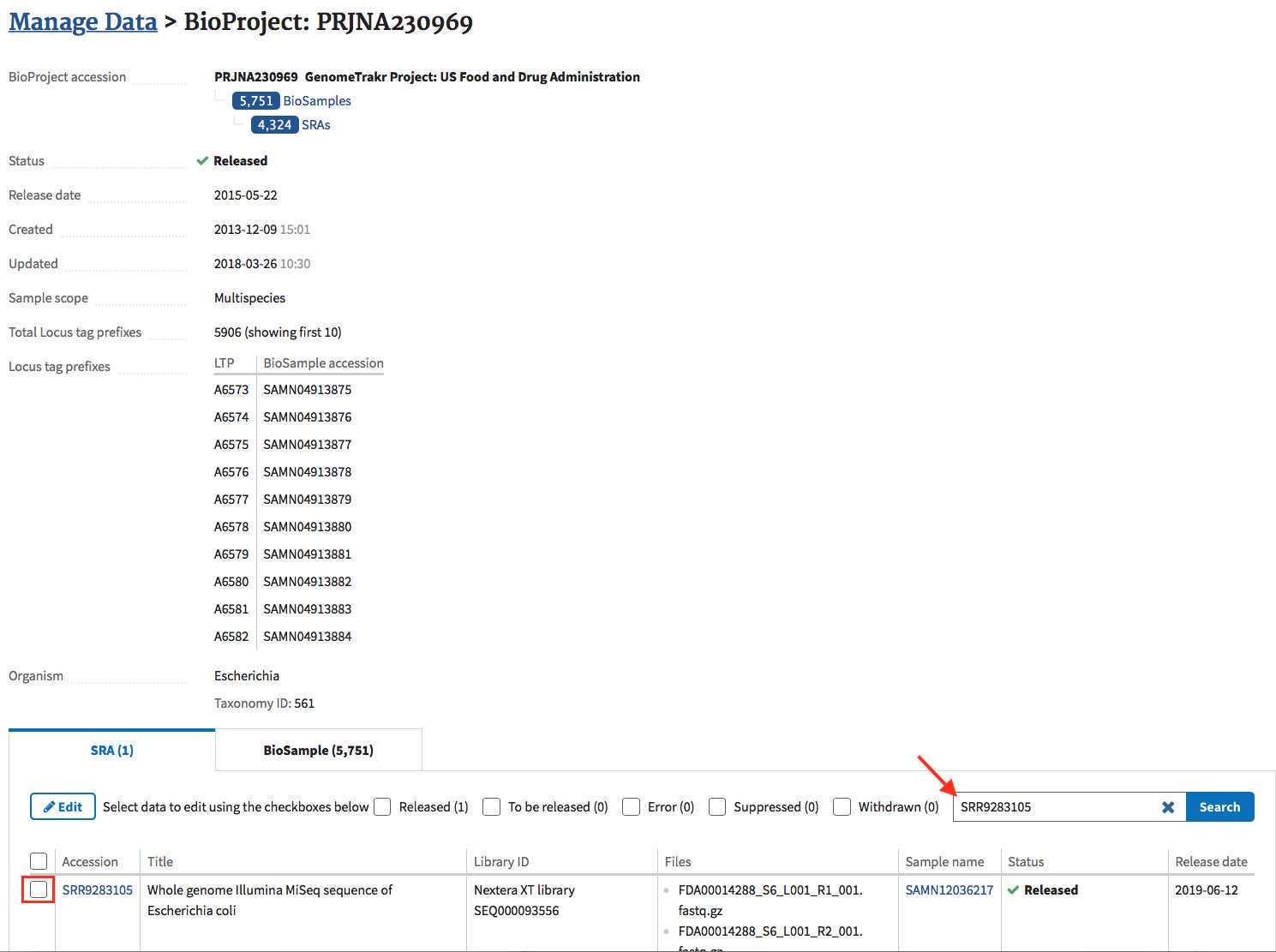
- You can now edit the metadata directly for this record. If you need to correct a sample-swap you can enter the correct BioSample accession here and the sequence will get re-parented.

Editing/updating custom SRA metadata attributes custom SRA metadata attributes
SRA inquiries : sra@ncbi.nlm.nih.gov
Attach a tab-delimited text file with the SRR accessions in the first column and attributes to update included as additional columns (*** only include columns you want to update*** ).
Examples:
FDA_SRA_update_20210203_ct.txt (adding custom wastewater attributes)
FDA_SRA_update_20210203_fb.txt (updating core SRA metadata attributes)
The following table will update or add the custom attributes used for the covid wastewater project:
| A | B | C | D | E |
|---|---|---|---|---|
| Run | enrichment_kit | amplicon_PCR_primer_scheme | library_preparation_kit | dehosting_method |
| SRR17540870 | NEBNext ARTIC SARS-CoV-2 RT-PCR Module | NEB VarSkip Short | Illumina DNA prep | SRA human read removal tool |
Tab-delimited table for updating an SRA record.
SRA retraction
Emails for SRA retraction: _sra@ncbi.nlm.nih.gov,_ sra@ncbi.nlm.nih.gov, pd-help@ncbi.nlm.nih.gov
*cc all retraction requests to PD-help, so they can ensure all linked records are retracted (GenBank, etc.).
An SRA record should only be retracted for the following reasons:
- Discovery of poor quality data. Lab intends to re-generate data (starting at appropriate wet-lab step, re-isolation, DNA extraction, library prep, or sequencing) and re-submit the data.
- Sample mix-ups that cannot be resolved by re-parenting or correcting the BioSamples. Lab intends to re-generate (starting at appropriate wet-lab step, re-isolation, DNA extraction, library prep, or sequencing) and re-submit the data.
- Discovery of multiple runs per isolate. Laboratory would like to have only one run per isolate in the system. No re-sequencing planned.
DO NOT retract an SRA submission, then attempt to re-submit the same files. This will get flagged as a duplicate within NCBI's validation check and and will be rejected.
Emails should include a list of SRR accessions to retract and reason for retraction (i.e. sample mix-up, quality of data, etc.).
Email template:

