Modified NEBNext® VarSkip Short SARS-CoV-2 Enrichment and library prep for Oxford Nanopore Technologies- adapted for wastewater samples
Padmini Ramachandran, Tamara Walsky, Amanda Windsor, Maria Hoffmann, Christopher Grim
Abstract
This protocol details methods for the preparation of SARS-CoV-2 sequencing library using VSS primers from library preparation from NEBNext® ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies®), NEB #E7660S/L 24/96 reactions adapted for wastewater samples.
Standard Protocol with PCR Bead Cleanup: This protocol includes a cleanup and normalization step for each sample after cDNA synthesis. Performing the cleanup and normalization step creates library pools where the reads for each library are more evenly distributed. These pools will likely achieve sufficient and equal coverage in less run time, but they take more hands-on time
This protocol also includes options for barcoding with dual indexing or single indexing of the samples.
Before start
Note: The amount of RNA required for detection depends on the abundance of the RNA of interest. In general, we recommend, using > 10 copies of the (SARS-CoV-2) viral genome as input. In addition, we recommend setting up a no template control reaction and all reactions are set-up in a hood .
The presence of carry-over products can interfere with sequencing accuracy, particularly for low copy targets. Therefore, it is important to carry out the appropriate no template control (NTC) reactions to demonstrate that positive reactions are meaningful.
Steps
Before you start
cDNA Synthesis
Gently mix 10 times by pipetting and spin down the LunaScript RT SuperMix reagents (contains primers). Prepare the cDNA synthesis reaction as described below:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| RNA Sample* | 8 µl |
| (lilac) LunaScript RT SuperMix | 2 µl |
| Total Volume | 10 µl |
*Up to 0.5 µg total RNA can be used in a 10 µl reaction.
Flick the tube or pipet up and down 10 times to mix followed by a quick spin.
For no template controls, mix the following components:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| (white) Nuclease-free Water | 8 µl |
| (lilac) LunaScript RT SuperMix | 2 µl |
| Total Volume | 10 µl |
Flick the tube or pipet up and down 10 times to mix followed by a quick spin.
Incubate reactions in a thermocycler with lid temperature at 105°C with the following steps:
| A | B | C | D |
|---|---|---|---|
| CYCLE STEP | TEMP | TIME | CYCLE |
| Primer Annealing | 25°C | 2 minutes | 1 |
| cDNA Synthesis | 55°C | 20 minutes | |
| Heat Inactivation | 95°C | 1 minute | |
| Hold | 4°C | ∞ |
Targeted cDNA Amplification
Gently mix Q5 Hot Start High Fidelity 2X master mix 10 times by pipetting and spin down reagents. Prepare the split pool amplification reactions as described below:
For Pool set A:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| cDNA (Step 6) | 4.5 µl |
| (lilac) Q5 Hot Start High-Fidelity 2X MM | 6.25 µl |
| NEBNext VSS SARS-CoV-2 Primer Mix 1 | 1.75 µl |
| Total Volume | 12.5 µl |
For Pool Set B:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| cDNA (Step 6) | 4.5 µl |
| (lilac) Q5 Hot Start High-Fidelity 2X MM | 6.25 µl |
| NEBNext VSS SARS-CoV-2 Primer Mix 2 | 1.75 µl |
| Total Volume | 12.5 µl |
Flick the tube or gently pipet up and down 10 times to mix followed by a quick spin.
Incubate reactions in a thermocycler* with the following steps:
| A | B | C | D |
|---|---|---|---|
| CYCLE STEP | TEMP | TIME | CYCLES |
| Initial Denaturation | 98°C | 30 seconds | 1 |
| Denature | 95°C | 15 seconds | 35 |
| Annealing/Extension | 63°C | 5 minutes | |
| Hold | 4°C | ∞ | 1 |
- Set heated lid to 105°C.
Cleanup of cDNA Amplicons
We highly recommend the clean up step using either NEBNext sample purification beads or Ampure beads.
For each sample, combine pool A and pool B PCR Reactions.
Vortex SPRIselect or NEBNext Sample Purification Beads to resuspend.
Add 20µL to the combined PCR reaction. Mix well by flicking the tube or pipetting up and down 10 times to mix and a very short 2-3 seconds quick centrifugation. Be sure to stop the centrifugation before the beads start to settle out.
Incubate samples at Room temperature for 0h 10m 0s.
Place the tubes on an appropriate magnetic stand to separate the beads from the supernatant. If necessary, quickly spin the sample 0h 0m 1s to collect the liquid from the sides of the tube before placing on the magnetic stand.
After 4 minutes (or when the solution is clear), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Add 500µL to the tube while in the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Repeat previous step once for a total of two washes:
Add 500µL to the tube while in the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Be sure to remove all visible liquid after the second wash. If necessary, briefly spin the tube for , place back on the magnetic stand and remove traces of ethanol with a p10 pipette tip. 0h 0m 1s, place back on the magnetic stand and remove traces of ethanol with a p10 pipette tip.
Air dry the beads for 0h 0m 30s while the tube is on the magnetic stand with the lid open.
Remove the tube from the magnetic stand. Elute the DNA target from the beads by adding 18µL.
Mix well by flicking the tube or pipetting up and down 10 times to mix and followed by a very short centrifugation. Incubate for 0h 10m 0s at Room temperature. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand.
Place the tube on the magnetic stand. After 2 minutes (or when the solution is clear), transfer 17µL to clean PCR tubes.
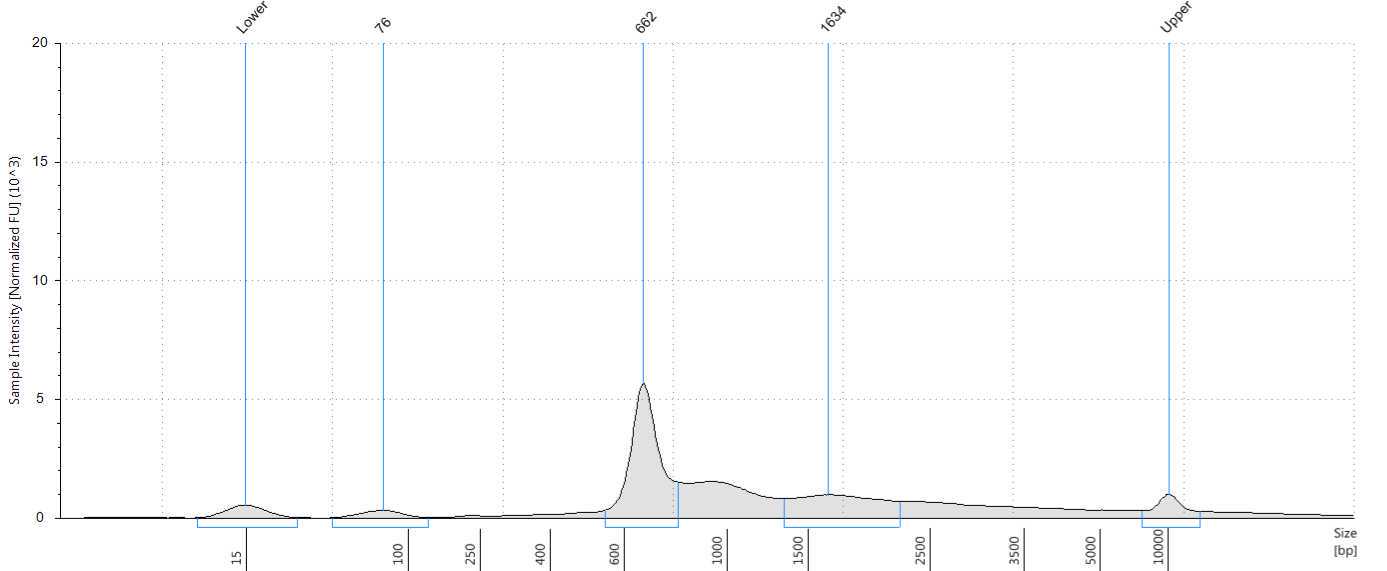
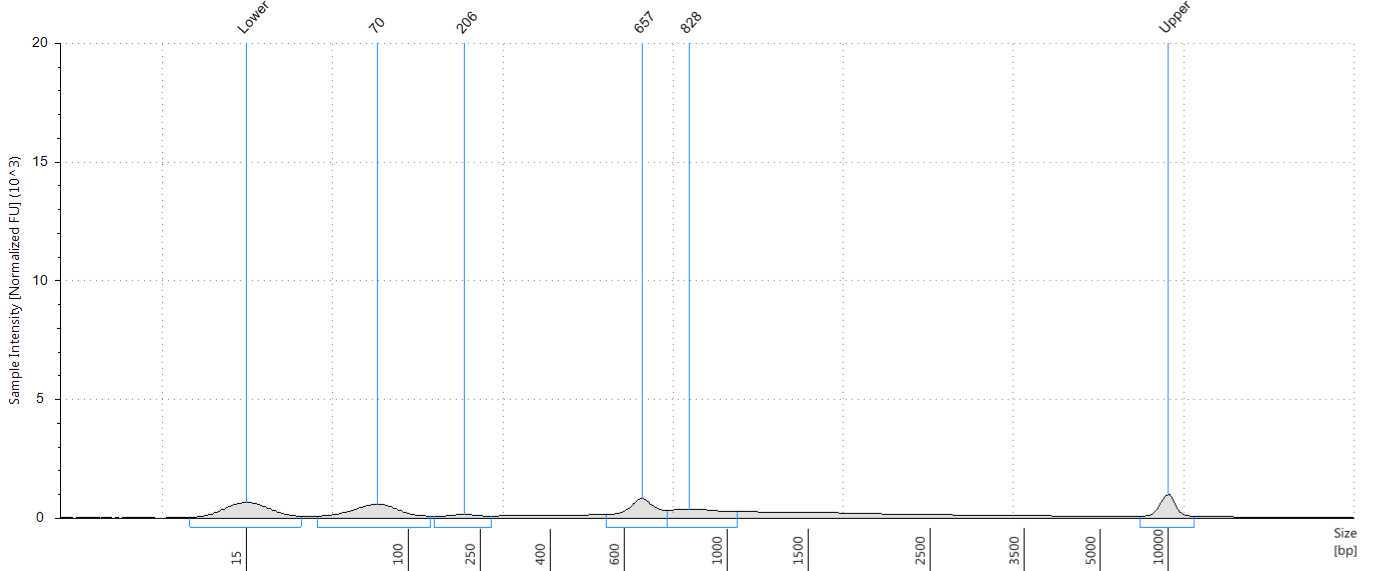
Assess the concentration of the DNA targets. We recommend using a Qubit fluorometer for concentration assessment. Use 1 µl of sample for the Qubit fluorometer. Amplicons may also be run on a Bioanalyzer® or Tape Station using High Sensitivity (HS) 5000 tape or HS 1000 tape to confirm ~560-650 bp size of amplicons.
NEBNext End Prep
Use the Qubit readings from Step 24 to determine the amount of the VSS Amplicons. Dilute each amplicon sample into 50 ng/12.5 μl (4ng/ul) concentration using Nuclease-free water. Add the following components to a PCR tube (End Prep Reaction and Buffer can be pre-mixed and stable On ice for 4 hours):
| A | B |
|---|---|
| COMPONENT | VOLUME |
| Targeted cDNA Amplicons (Step 24) | 12.5 µl |
| (green) NEBNext Ultra II End Prep Reaction Buffer | 1.75 µl |
| (green) NEBNext Ultra II End Prep Enzyme Mix | 0.75 µl |
| Total Volume | 15 µl |
Flick the tube or gently pipet up and down 10 times to mix the solution. Perform a quick spin to collect all liquid from the sides of the tube.
Place in a thermocycler, with the heated lid set to ≥ 75°C, and run the following program:
0h 10m 0s @ 20°C
0h 10m 0s @ 65°C
Hold at 4°C
Barcode Ligation
Add the following components directly to a sterile nuclease-free PCR tube:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| End-prepped DNA (Previous Step) | 3 ul |
| Dual Barcode* | 8 ul |
| (red) Blunt/TA Ligase Master Mix** | 10 µl |
| Total Volume | 21 µl |
- Barcodes are provided in Oxford Nanopore Technologies Dual Barcoding Expansion kit EXP- NBD 196. Adding 8ul of barcode to 3-5ul of End prepped DNA helps with better barcoding efficiency for wastewater samples.** Mix the Blunt/TA Ligase Master Mix by pipetting up and down several times prior to adding to the reaction.
Flick the tube or gently pipet up and down 10 times to mix solution. Perform a quick spin to collect all liquid from the sides of the tube.
Incubate at 20Room temperature for 0h 20m 0s
Incubate at 65°C for 0h 10m 0s.
Place 65On ice for 0h 1m 0s.
Pool all barcoded samples into one 1.5 ml DNA LoBind Tube.
Cleanup of Barcoded DNA
The following section is for cleanup of the ligation reaction.
Vortex NEBNext Sample Purification Beads to resuspend.
Add 0.4X resuspended beads to pooled, barcoded samples (Step 30), for example, if you are pooling 12 samples with 2 barcode set up, which will be 24 libraries (which amounts to 480 µl total), add 192µLto the 480 µl of pooled sample. Flick the tube or pipet up and down 10 times to mix to resuspend pellet. Perform a quick spin for 0h 0m 1s to collect all liquid from the sides of the tube.
Incubate samples on bench top for 0h 10m 0s at 65Room temperature.
Place the tube on a 1.5 ml magnetic stand (such as NEB S1506) to separate the beads from the supernatant. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing on the magnetic stand.
After 5 minutes (or when the solution is clear), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Wash the beads by adding 250µL. Flick the tube or pipet up and down to mix to resuspend pellet. If necessary, quickly spin the sample for 0h 0m 1s to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand.
Place the tube on an appropriate magnetic stand for 4 minutes (or until the solution is clear) to separate the beads from the supernatant. Remove the supernatant.
Repeat previous 2 steps once for a total of two washes:
Wash the beads by adding 250µL. Flick the tube or pipet up and down to mix to resuspend pellet. If necessary, quickly spin the sample for 0h 0m 3s to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand.
Place the tube on an appropriate magnetic stand for 4 minutes (or when the solution is clear) to separate the beads from the supernatant. Remove the supernatant.
Be sure to remove all visible liquid after the second wash. If necessary, briefly spin the tube, place back on the magnetic stand and remove traces of SFB with a p10 pipette tip
Add 200µL to the tube while on the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Perform a quick spin and place the sample tube on the magnetic stand, to remove any residual ethanol.
Air dry the beads for 0h 0m 30s while the tube is on the magnetic stand with the lid open.
Remove the tube from the magnetic stand. Elute the DNA target from the beads by adding 33µL.
Resuspend the pellet by flicking the tube or pipetting up 10 times and down to mix. Incubate for at least 2 minutes at Room temperature. If necessary, quickly spin the sample for 0h 0m 1s to collect the liquid from the sides of the tube before placing back on the magnetic stand.
Place the tube on the magnetic stand. After 2 minutes (or when the solution is clear), transfer 32µL to a new 1.5 ml microcentrifuge DNA LoBind Tube or PCR tube.
We recommend assessing cDNA concentrations with a Qubit fluorometer. Use 1 µl for the Qubit fluorometer .
Adapter Ligation
Add the following components into a 1.5 ml microcentrifuge DNA LoBind Tube or nuclease-free PCR tube:
| A | B |
|---|---|
| COMPONENT | VOLUME |
| Dual barcoded and purified DNA (Step 45) | 30 µl |
| Adapter Mix II (AMII)** | 5 µl |
| (red) NEBNext Quick Ligation Reaction Buffer * | 10 µl |
| (red) NEBNext Quick T4 Ligase | 5 µl |
| Total Volume | 50 µl |
- Mix the NEBNext Quick Ligation Reaction Buffer by pipetting up and down several times prior to adding to the reaction. ** Adapter Mix II is provided by Oxford Nanopore Technologies Native Barcoding Expansion 1-12 (EXP-NBD104), 13-24 (EXP-NBD114) and 1-96 (EXP-NBD-196) kits.
Flick the tube to mix solution. Perform a quick spin for 0h 0m 1s to collect all liquid from the sides of the tube.
Incubate at 25°C or at 20Room temperature for 0h 20m 0s.
Proceed to Cleanup of Adapter-ligated DNA in the next section.
Cleanup of Adapter Ligated DNA
Vortex NEBNext Sample Purification Beads to resuspend.
Add 40µL to the ligation mix. Mix well by flicking the tube to mix followed by a quick spin for 0h 0m 1s.
Incubate samples for 0h 15m 0s at 20Room temperature.
Place the tube on an appropriate magnetic stand to separate the beads from the supernatant. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing on the magnetic stand.
After 5 minutes (or when the solution is clear), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA libraries.
Wash the beads by adding 250µL. Flick the tube to mix to resuspend pellet. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand. Place the tube on an appropriate magnetic stand.
Wait for 5 minutes (or until the solution is clear) to separate the beads from the supernatant. Remove the supernatant.
Repeat previous 2 steps once for a total of two washes:
Wash the beads by adding 250µL. Flick the tube to resuspend pellet. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand. Place the tube on an appropriate magnetic stand.
Wait for 5 minutes (or when the solution is clear) to separate the beads from the supernatant. Remove the supernatant.
Be sure to remove all visible liquid after the second wash. If necessary, briefly spin the tube/plate, place back on the magnet and remove traces of SFB with a p10 pipette tip.
Remove the tube from the magnetic stand. Elute the DNA target from the beads by adding 15µL provided in SQK-LSK109 kit from Oxford Nanopore.
Resuspend the pellet well in EB buffer by flicking the tube. Incubate for 0h 15m 0s at 20Room temperature. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand.
Place the tube/plate on the magnetic stand. After 5 minutes (or when the solution is clear), transfer 15µL to a new DNA LoBind tube.
Use Qubit to quantify 1µL. Follow Oxford Nanopore Protocol SQK-LSK109 to prepare MinION® flow cell and DNA library sequencing mix and load the flow cell. We recommend not multiplexing more than 10 samples(9 samples + NTC) in one R9.4.1 flow cell. We highly recommend using a Negative template control and label them as 'water', 'negative', 'blank', 'ntc' if using our custom analysis pipeline for the data analysis.
The base calling options can be one among the three: Fast base calling, High accuracy base calling, or Super accurate base calling. We have observed improved average read quality with High accuracy and Super accurate base calling, but little difference in read numbers, or variant calling compared to Fast base calling, using our custom analysis pipeline.