Modified NEBNext® VarSkip Long SARS-CoV-2 Enrichment and library prep (Native Barcoding Kit V14 Oxford Nanopore Technologies)- adapted for wastewater samples
Padmini Ramachandran, Tamara Walsky, Amanda Windsor, Maria Hoffmann, Christopher Grim, Kathryn Judy
Disclaimer
Abstract
This protocol details methods for the preparation of SARS-CoV-2 sequencing library using VSL primers from NEB, adapted for wastewater samples. This protocol produces multiplexed amplicon libraries suitable for sequencing on Oxford Nanopore Technologies® (ONT) MinION systems using ONT V14 chemistry (SQK-NBD114).
Before start
Note: We recommend setting up a no template control reaction and all reactions are set-up in a biological safety cabinet .
The presence of carry-over products can interfere with sequencing accuracy, particularly for low copy targets. Therefore, it is important to carry out the appropriate no template control (NTC) reactions to demonstrate that positive reactions are meaningful.
Steps
Before you start
Targeted cDNA Amplification
Prepare master mixes fresh immediately before performing cDNA amplification.
- Q5 Hot Start High-Fidelity Polymerase should stay on ice at all times. Do not vortex.
- Thaw Q5 Reaction Buffer, MgCl2, dNTPs, and water.
- Mix thawed tubes, spin down, and place on ice.
- Thaw VarSkip Long Primer Mix 1 and VarSkip Long Primer Mix 2.
- Mix by flicking and spin down both the tubes.
- Keep on ice.
Prepare the split pool amplification reactions as described below:
For Pool set A:
Prepare the master mix below in sufficient volume for your samples.
| A | B |
|---|---|
| COMPONENT | VOLUME |
| Q5 Reaction Buffer | 2.5 µl |
| 50mM Magnesium Chloride | 0.5 µl |
| Deoxynucleotide (dNTP) Solution | 0.75 µl |
| Nuclease-free water | 1.75 µl |
| NEBNext VarSkip Long Primer Mix 1 | 2.25 µl |
| Total Volume | 7.5 µl |
For Pool Set B:
Prepare the master mix below in sufficient volume for your samples.
| A | B |
|---|---|
| COMPONENT | VOLUME |
| Q5 Reaction Buffer | 2.5 µl |
| 50mM Magnesium Chloride | 0.5 µl |
| Deoxynucleotide (dNTP) Solution | 0.75 µl |
| Nuclease-free water | 1.75 µl |
| NEBNext VarSkip Long Primer Mix 2 | 2.25 µl |
| Total Volume | 7.5 µl |
Mix the two master mix tubes by flicking and spin down. Dispense 7.5 µl master mix from each tube into separate PCR tube strips ( A and B ), two PCR tubes (one for each master mix) per sample to amplify.
Add 4.5 µl cDNA into each pre-filled PCR tube, ensuring each sample to be amplified is added into exactly 1 tube in strip A and 1 tube in strip B.
While keeping the polymerase on ice, add 0.5µL Q5 Hot Start High-Fidelity Polymerase to each tube.
Gently flick the tube strips to mix and spin down briefly.
Incubate Pool A reactions in a thermocycler* with the following steps:
| A | B | C | D |
|---|---|---|---|
| CYCLE STEP | TEMP | TIME | CYCLES |
| Initial Denaturation | 98°C | 30 seconds | 1 |
| Denature | 95°C | 15 seconds | 38 |
| Annealing | 59°C | 1 minute | |
| Extension | 72°C | 2 minutes | |
| Hold | 4°C | ∞ | 1 |
- Set heated lid to 105°C.
Incubate Pool B reactions in a thermocycler* with the following steps:
| A | B | C | D |
|---|---|---|---|
| CYCLE STEP | TEMP | TIME | CYCLES |
| Initial Denaturation | 98°C | 30 seconds | 1 |
| Denature | 95°C | 15 seconds | 38 |
| Annealing | 61°C | 45 seconds | |
| Extension | 72°C | 2 minutes | |
| Hold | 4°C | ∞ | 1 |
- Set heated lid to 105°C.
Cleanup of cDNA Amplicons
We highly recommend this clean up step using AMPure® XP beads, though NEBNext sample purification beads can be used as well.
For each sample, combine pool A and pool B PCR products (amplicons), measuring the pooled volume.
Vortex AMPure® XP beads for 0h 0m 30s to resuspend.
Add 0.6X to the combined PCR product. Mix well by flicking the tube and a very short 2-3 seconds quick centrifugation. Be sure to stop the centrifugation before the beads start to settle out.
Incubate samples at Room temperature for 0h 5m 0s.
Quickly spin samples to collect the liquid from the sides of the tube before placing on the magnetic stand for 0h 5m 0s to separate the beads from the supernatant.
After 5 minutes (or when the solution is clear), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Add 200µL to the tube while in the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Repeat previous step once for a total of two washes:
Add 200µL to the tube while in the magnetic stand. Incubate at Room temperature for 0h 0m 30s, and then carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
Be sure to remove all visible liquid after the second wash. If necessary, briefly spin the tube for , place back on the magnetic stand and remove traces of ethanol with a p10 pipette tip. 0h 0m 1s, place back on the magnetic stand and remove traces of ethanol with a p10 pipette tip.
Air dry the beads for up to 0h 3m 0s while the tube is on the magnetic stand with the lid open.
Remove the tube from the magnetic stand. Elute the DNA target from the beads by adding 18µL.
Mix well by flicking the tube followed by a very short centrifugation. Incubate for 0h 5m 0s at Room temperature. If necessary, quickly spin the sample to collect the liquid from the sides of the tube or plate wells before placing back on the magnetic stand.
Place the tube on the magnetic stand. After 0h 2m 0s (or when the solution is clear), transfer 17µL to clean PCR tubes.
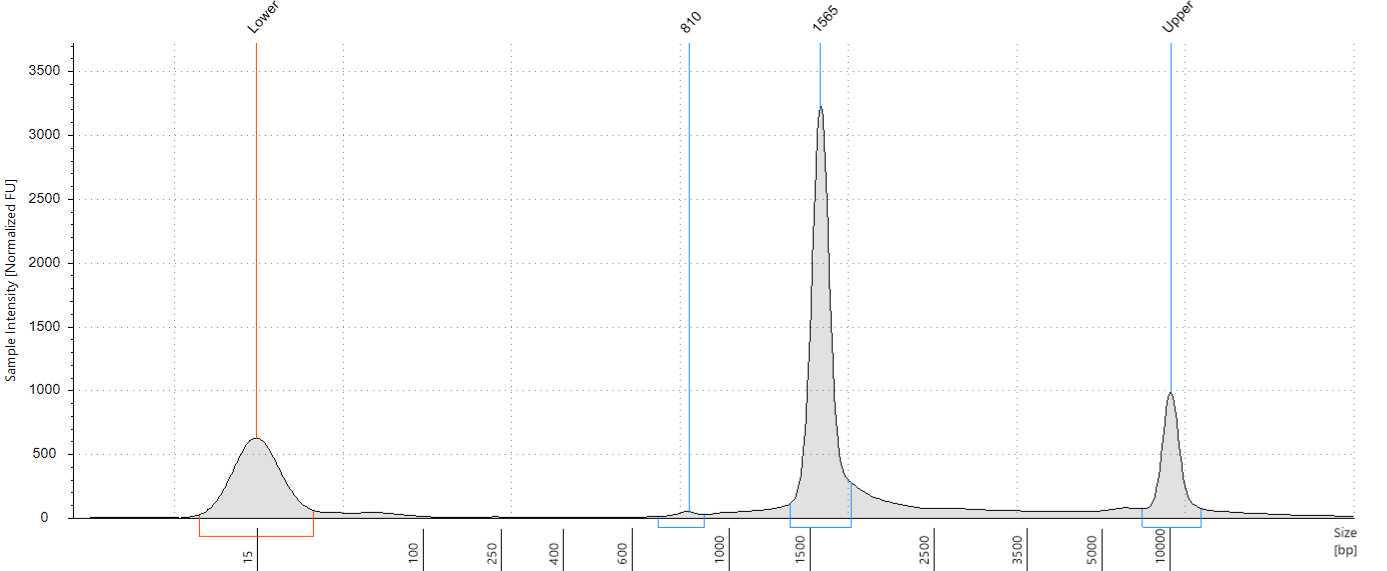
Assess the concentration of the DNA targets. We recommend using a Qubit fluorometer for concentration assessment. Use 1 µl of sample for the Qubit fluorometer. Amplicons should also be run on Femto or Bioanalyzer® or Tape Station using High Sensitivity (HS) 5000 tape to confirm ~1500-1600 bp size of amplicons.
End-Prep
Calculate the volume of each sample needed to bring forward at least 200fmol per sample using the amplicon size determined after cleanup. We recommend bringing forward approximately the same moles of DNA for each sample.
Aliquot the volume of each sample calculated into fresh PCR tubes and make up each sample to 11.5µL using nuclease-free. Excess amplicons should be returned to 4°C
Make the end repair and dA-tailing master mix by combining the reagents below in the order and amounts listed in the table. Adjust component volumes for your number of samples plus 20% overage.
- Thaw DNA Control Sample (DCS) at room temperature, vortex briefly, and place on ice. If this is the first use of the DCS tube, dilute by adding
105µL, mix gently by pipetting, and spin down. Diluted DCS can be stored at-20°Cafter use in Step 24. - Thaw Ultra II End-Prep Buffer at room temperature, then vortex and spin down briefly.
- Thaw Ultra II End Prep Enzyme mix on ice, spin down briefly, and return to ice. Do not vortex.
| A | B |
|---|---|
| Component | Volume per Sample |
| Ultra II End-prep reaction buffer | 1.75 µl |
| Ultra II End-prep enzyme mix | 0.75 µl |
| Total volume | 2.5 µl |
Mix the master mix components by pipetting or gentle flicking and quickly centrifuge. Master mix can remain stable On ice for 4 hours.
Add 1µL to each sample, mix by gentle flicking, and spin down briefly.
Add 2.5µL to each sample, mix by gently flicking the tubes, and spin down briefly.
Incubate samples in a thermocycler* with the following settings:
| A | B | C | D |
|---|---|---|---|
| TEMP | TIME | CYCLES | |
| 20°C | 5 minutes | 1 | |
| 65°C | 5 minutes | 1 | |
| 4°C | ∞ | 1 |
- Set heated lid to 105°C
Native Barcode Ligation and Cleanup
In PCR tubes or a 96-well plate, add reagents below in the order listed in the table.
- Thaw Blunt/TA Ligase master mix at room temperature, spin down 5 seconds, then mix with 10 full volume pipette mixes and place on ice. Do not vortex.
- Thaw EDTA at room temperature, mix by vortexing, spin down, and place on ice.
- Thaw Native Barcodes (ex: NB01-96) required for your number of samples at room temperature, individually mix by pipetting, spin down, and place on ice.
- Add a unique barcode to each sample to be run together on a single flowcell (to be pooled).
- 2-3 barcoding reactions per sample may be desired to increase input DNA. If multiple reactions per sample are performed, be sure to use the same barcode for the same sample in each reaction.
| A | B |
|---|---|
| Component | Volume per Sample |
| End-prepped DNA | 3 µl |
| Native Barcode | 5 µl |
| Blunt/TA Ligase master mix | 5 µl |
| Total | 13 µl |
Mix components by gently flicking the tubes, then centrifuge briefly.
Incubate samples at Room temperature for 0h 20m 0s
Add 1µL to each tube to stop the reaction. Mix well by flicking and spin down briefly.
Pool all barcoded samples to be run on a single flowcell in a 1.5 µl LoBind tube, measuring the volume each sample as it is added. Calculate the final volume of the pooled samples.
Add 0.4X to each pool and mix by gently flicking followed by a short spin to collect the liquid. Stop the centrifugation before the beads begin to settle.
Incubate at room temperature for 0h 10m 0s to bind DNA to the beads, agitating the pool(s) every two minutes. If available, pools can be incubated on a Hula mixer (rotator mixer) to agitate the beads instead.
Spin down the pool(s) and place tube(s) in an appropriate magnetic separation rack until the beads have separated, 0h 5m 0s
Carefully pipette off the supernatant without disturbing the beads, discarding the supernatant. Do not discard the beads, which contain your DNA target .
Wash the beads with 700µL. Pipette off the ethanol and discard. If the pellet was disturbed, wait for the beads to pellet again before pipetting off the ethanol. Do not discard the beads.
Repeat the previous step once for a total of two washes:
Wash the beads with 700µL. Pipette off the ethanol and discard. If the pellet was disturbed, wait for the beads to pellet again before pipetting off the ethanol. Do not discard the beads.
To remove residual ethanol, quickly spin pool(s) and return the tube(s) to the magnetic rack, allowing beads to separate fully. Pipette off residual ethanol with a P20 pipette and discard. Do not discard the beads.
Remove samples from the magnetic rack and immediately add 35µL. Resuspend beads by flicking, then quickly spin to collect liquid.
Incubate pool(s) at 37°C for 0h 10m 0s to elute DNA. Every two minutes, agitate the sample by gentle flicking for 10 seconds to encourage elution.
Place samples on the magnetic rack until beads separate fully from the solution, 0h 5m 0s
Slowly pipette 35µL of clear eluate without disturbing the beads and transfer to new 1.5 µl LoBind tube(s). Discard the old sample tube(s) with beads. Do not discard the supernatant.
Adapter Ligation and Cleanup
- Thaw NEBNext Quick Ligation Reaction Buffer at room temperature, pipette up and down several times to break up precipitate, vortex for several seconds, then spin down briefly.
- Spin down Quick T4 DNA Ligase, pipette mix, and place on ice. Do not vortex.
- Spin down Native Adapter, pipette mix, and place on ice.
- Thaw Short Fragment Buffer (SFB) at room temperature, mix by vortexing, spin down, and place on ice.
| A | B |
|---|---|
| Component | Volume per Sample |
| Pooled barcoded sample | 30 µl |
| Native Adapter | 5 µl |
| NEBNext Quick Ligation Reaction Buffer | 10 µl |
| Quick T4 DNA Ligase | 5 µl |
| Total volume | 50 µl |
Thoroughly mix components by pipetting or gently flicking the tube, then quickly centrifuge to mix.
Incubate the reaction for 0h 20m 0s at room temperature.
Thaw elution buffer at room temperature.
Add 20µL (0.4X) to each nuclease-treated sample and mix by gently flicking followed by a short spin to collect the liquid. Stop the centrifugation before the beads begin to settle.
Incubate at room temperature for 0h 10m 0s to bind DNA to the beads, agitating every two minutes. If available, the library can be incubated on a Hula mixer (rotator mixer) to agitate the beads instead.
Spin down and place tube in an appropriate magnetic separation rack until the beads have separated, 0h 5m 0s
Carefully pipette off the supernatant without disturbing the beads, discarding the supernatant. Do not discard the beads, which contain your DNA target .
Add 125µL to each sample tube. Flick the tube to resuspend beads, collect liquid with a quick spin, and return the tube to the magnetic rack.
When the beads have pelleted, remove the supernatant and discard without disturbing the beads. Do not discard the beads.
Repeat the previous step once for a total of two washes:
Add 125µL to each sample tube. Flick the tube to resuspend beads, collect liquid with a quick spin, and return the tube to the magnetic rack.
When the beads have pelleted, remove the supernatant and discard without disturbing the beads. Do not discard the beads.
To remove residual Short Fragment Buffer, quickly spin the tube and return to the magnetic rack, allowing beads to separate fully. Pipette off residual supernatant with a P20 pipette and discard. Do not discard the beads.
Remove the tube from the magnetic rack and immediately add 15µL. Resuspend beads by flicking, then quickly spin to collect liquid.
Incubate at 37°C for 0h 10m 0s to elute DNA. Every two minutes, agitate the sample by gentle flicking for 10 seconds to encourage elution.
Place tube on the magnetic rack until beads separate fully from the eluate, usually less than 0h 5m 0s
Slowly pipette 15µL of clear supernatant without disturbing the beads and transfer to a new LoBind tube. Discard the old tube with beads. Do not discard the supernatant.
Measure DNA concentration with a Qubit Fluorometer using the 1x dsDNA HS kit. The final library should also be run on Femto or Bioanalyzer® or Tape Station using High Sensitivity (HS) 5000 tape to confirm the size of the final library.
End Protocol
Based on the concentration and library size determined in step 55, aliquot 10-20fmol and make up to 12µL using nuclease-free water in a new 0.5 µl LoBind tube. This will be used to load the flowcell.
Prime and load the R10.4.1 (FLO-MIN114) SpotON flowcell following the Oxford Nanopore SQK-LSK114 protocol. We recommend using High Accuracy Basecalling (HAC) at 260bps ("Accurate") speed.