Library clean up and quality control for Illumina sequencing
Katherine Smollett, Lily Tong, Jenna Nichols, Kirsty Kwok, Kyriaki Nomikou, Ma. Jowina Galarion, Daniel Mair, Ana Filipe
Disclaimer
This work is licensed under a Creative Commons Attribution 4.0 International License.

Abstract
There are many different ways of preparing libraries for Illumina sequencing but when it comes to the post-library preparation clean up and quality control (QC) there are common rules to apply. For reliable sequencing results it is essential that certain criteria are reached and so accurate QC is crucial. Here we describe a standard library clean up and QC with tips for how to rectify common problems including low yield, adapter dimers and daisy chains.
Before start
Library clean-ups should be done in a separate lab or area to the pre-PCR stage of the library preparation to prevent contamination.
Steps
Library clean up
Equilibrate Ampure XP beads to Room temperature for 0h 30m 0s and vortex well to mix.
If needed make libraries up to 50µL with 10mM Tris pH8.
Add 45µL to the samples (ratio 0.9:1) and mix well.
Incubate at Room temperature room temperature for 0h 5m 0s.
Place on a magnetic rack for 0h 5m 0s until beads and solution have fully separated.
Carefully remove supernatant without disturbing the beads.
Carefully add 200µL without disturbing the beads.
Incubate at Room temperature for 0h 0m 30s.
Place on magnetic rack for 0h 1m 0s until the beads and solution have fully separated.
Keeping on the magnet and carefully remove supernatant without disturbing the beads.
Repeat wash with 200µL.
Remove all traces of Ethanol.
Keeping on the magnet air dry for up to 0h 3m 0s.

Add 18µL, remove from the magnet and mix well to fully suspend the beads.
Incubate at Room temperature for at least 0h 2m 0s to elute the DNA.
Place back on magnet until the beads and solution have fully separated.
Transfer the supernatant containing the library DNA to a fresh tube.
Library quantification
It is recommended that library quantification is performed using a fluorometric method (e.g. Qubit) rather than an absorbance based method (e.g. Nanodrop) as it is more specific, sensitive and accurate.
Equipment
| Value | Label |
|---|---|
| Qubit | NAME |
| Flurometer | TYPE |
| Invitrogen | BRAND |
| Q33228 | SKU |
Prepare a 1:5 dilution of each sample by adding 2µL to 8µL in fresh tubes.
Prepare 0.5 mL thin-walled PCR tubes, including 2 tubes for the standard solution and 1 tube per sample. Label the tube lids and not the sides.
Prepare the Qubit dsDNA High Sensitivity master mix for the total number of samples and standards with an excess as follows:
| A | B |
|---|---|
| Component | Volume (μl) |
| Qubit dsDNA HS Buffer | 199 |
| Qubit dsDNA HS Reagent | 1 |
| Total | 200 |
Aliquot the Qubit dsDNA High Sensitivity master mix into the assay tubes as follows:
Standards 190µL
Samples 199µL
Add 10µL to each standard assay tube.
Add 1µL to each sample assay tube.
Vortex assay tubes and briefly centrifuge.
Incubate at Room temperature for 0h 2m 0s.
Select dsDNA high Sensitivity assay on the Qubit Fluorimeter and press "Read Standards".
Insert Standard 1 and 2 into the sample chamber when prompted, close the lid and and press "Read Standard".
Select "Run samples" and select the sample volume as 1µL.
Insert a sample tube into the sample chamber, close the lid and press "Read tube".
Record the concentration of the Qubit sample in ng/μL.
Calculate the concentration of the original sample by multiplying the Qubit concentration (see Step 31) by the dilution factor (see Step 19).
Library visualisation
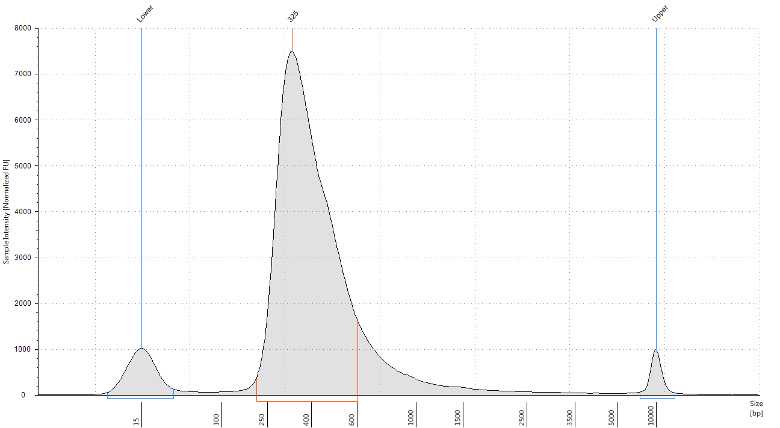
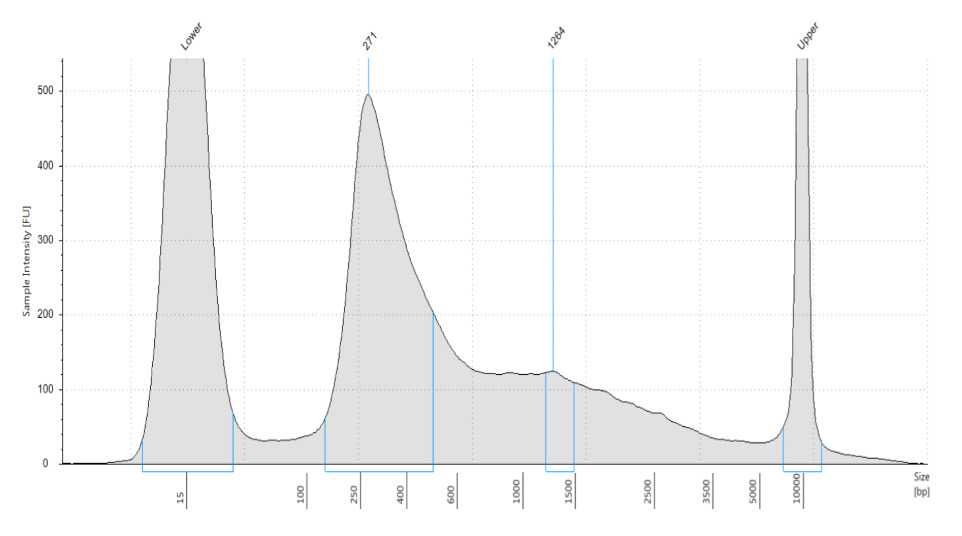
It is recommended that the libraries are visualised using capillary electrophoresis, we describe visualisation with a TapeStation and High Sensitivity D5000 ScreenTape. Alternatives such as the BioAnalyzer or Fragent Analyzer can also be used.
The purpose is to provide a size for the library fragments to give accurate molar quantification and to determine the quality of the library.
Equipment
| Value | Label |
|---|---|
| 4200 TapeStation System | NAME |
| Electrophoresis tool for DNA and RNA sample quality control. | TYPE |
| TapeStation Instruments | BRAND |
| G2991AA | SKU |
Ensure that the D5000 ScreenTape and Reagents are equilibrated to Room temperature for at least 0h 30m 0s before use, vortex and briefly centrifuge.
Dilute each sample to approximately 1 ng/μL.
In fresh PCR strip tubes prepare the ladder assay tube as follows:
| A | B |
|---|---|
| Component | Volume (μl) |
| D5000 sample buffer | 2 |
| D5000 ladder | 2 |
| Total | 4 |
Prepare the sample assay tubes as follows:
| A | B |
|---|---|
| Component | Volume (μl) |
| D5000 sample buffer | 2 |
| Diluted library | 2 |
| Total | 4 |
Spin down, using IKA vortexer mix at 2000rpm then spin down again.
Load the assay tubes and ScreenTape into the TapeStation instrument.
Select the required sample/ladder positions in the TapeStation software and click "start".
Determine the library peak size in bp.
Calculate the library molar concentration using the fragment size (Step 42) and sample mass concentration determined by Qubit (Step 32).
Library troubleshooting
Below we list a number of common problems associated with Illumina sequencing libraries and steps you can take to rectify them.
Low yield
If your library yield is very low (e.g. less than 1 ng/μL) then the library can be amplified using the KAPA HiFi Library Amplification kit which uses primers directed against the Illumina adapter sequences.
Prepare the PCR mix (for multiple reactions a master mix with 10% excess is recommended):
| A | B |
|---|---|
| Component | Volume (μl) |
| 2X KAPA HiFi Ready Mix | 25 |
| 10X Universal primer mix | 5 |
| Total | 30 |
Add 30µL to fresh PCR tubes.
Add 20µL.
Incubate as follows on PCR machine:
98°C for 0h 0m 45s
1-6 cycles of
98°C for 0h 0m 15s
65°C for 0h 0m 30s
72°C for 0h 0m 30s
Final cycle of
72°C for 0h 1m 0s
4°C .
Repeat library clean up and QC (Steps 1-43).
Adapter dimers
Adapter dimers are small fragments (typically ~120-170 bp) that contain full length adapter sequences meaning that they are able to bind to the Illumina flow cell and be clustered. Due to the small size they cluster more efficiently than the full-length libraries meaning that it is very important that they are removed before sequencing.
Adapter dimers cause a number of problems:
- Reduce the number of library specific reads.
- Over clustering, reducing the data quality for all the samples.
- Shorter length than run length results in adapter dimer cluster giving no signal in later run cycles reducing quality.
- Free adapter can be incorporated into library clusters causing index hopping.
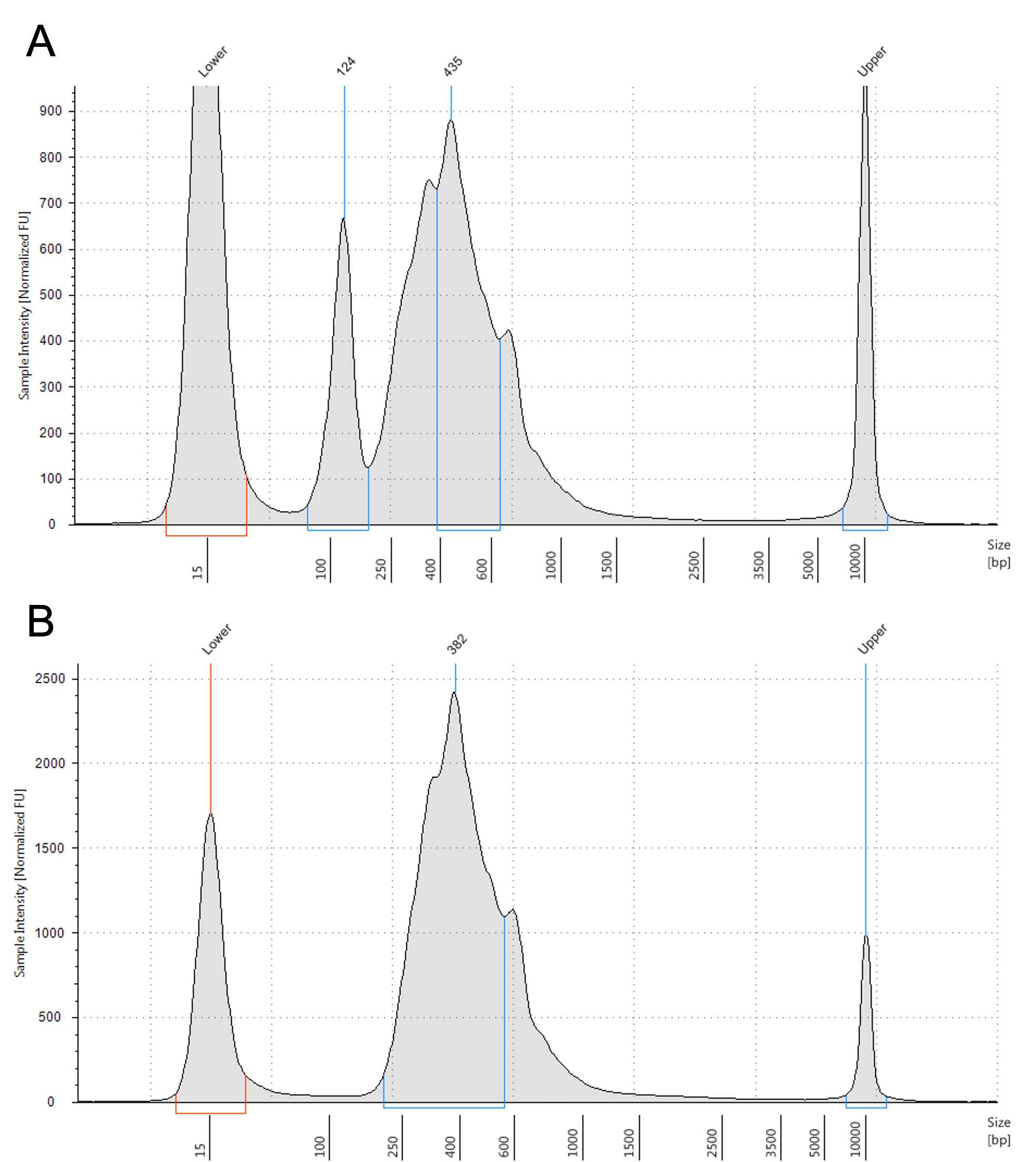
Adapter dimers are removed by repeating the Ampure XP clean up (Steps 1-17)
If your specific library fragment size permits a more stringent ratio of Ampure:sample (e.g. 0.8X or 0.7X) can be used.
Depending on the extent of the adapter dimer, multiple rounds of clean up can be performed.
After you have completed your rounds of clean up repeat the QC (Steps 18-43)
Daisy chains
Assemblies of improperly annealed, partially double-stranded, hetero-duplex DNA called daisy chains can result from over amplification causing the depletion of primers/dNTP's in later cycles, or contaminants such as ethanol carried over from incomplete Ampure bead clean up into the final PCR reaction.
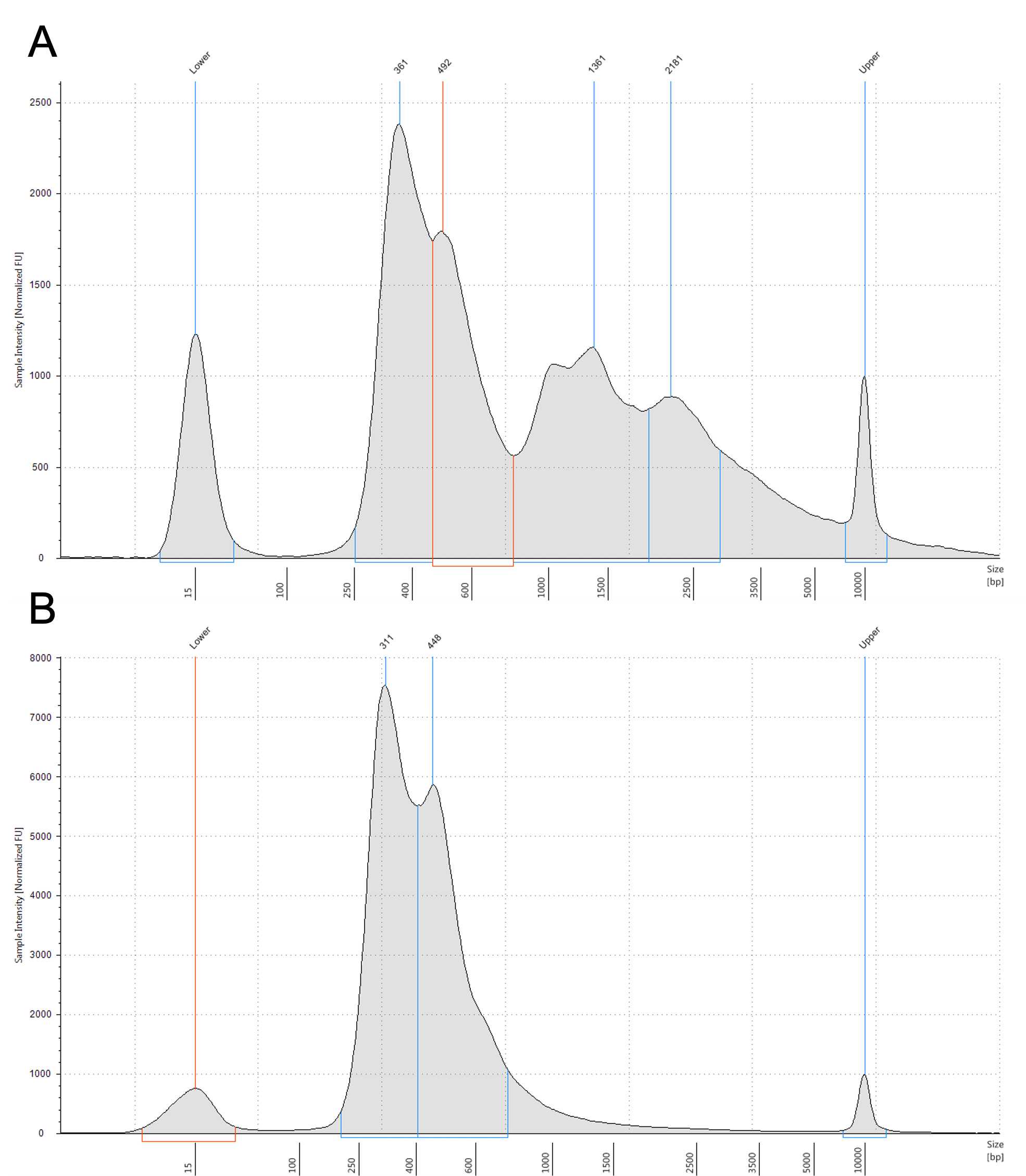
Daisy chains can be broken up by performing 1-2 additional PCR cycles (Steps 45.1 - 45.5).
Large fragment contamination
Contamination with larger fragments are less serious than small fragment contamination and adapter dimers, as they cluster less efficiently than the smaller, specific libraries. They do interfere with accurate quantification and can be removed by reverse-ampure clean up.
If needed make libraries up to 50µL with 10mM Tris pH8.
Add 30µL to the samples (ratio 0.6:1).
Incubate at Room temperature room temperature for 0h 5m 0s.
Place on a magnetic rack for 0h 5m 0s until beads and solution have fully separated.
Carefully transfer the supernatant containing the smaller DNA fragments to fresh tubes.
Recover the DNA fragments form the supernatant by adding 60µL (including the solution from step 48.2 this makes a ratio 1.8:1).
Incubate at Room temperature room temperature for 0h 5m 0s.
Continue with library clean up and QC as above (Steps 7-43).