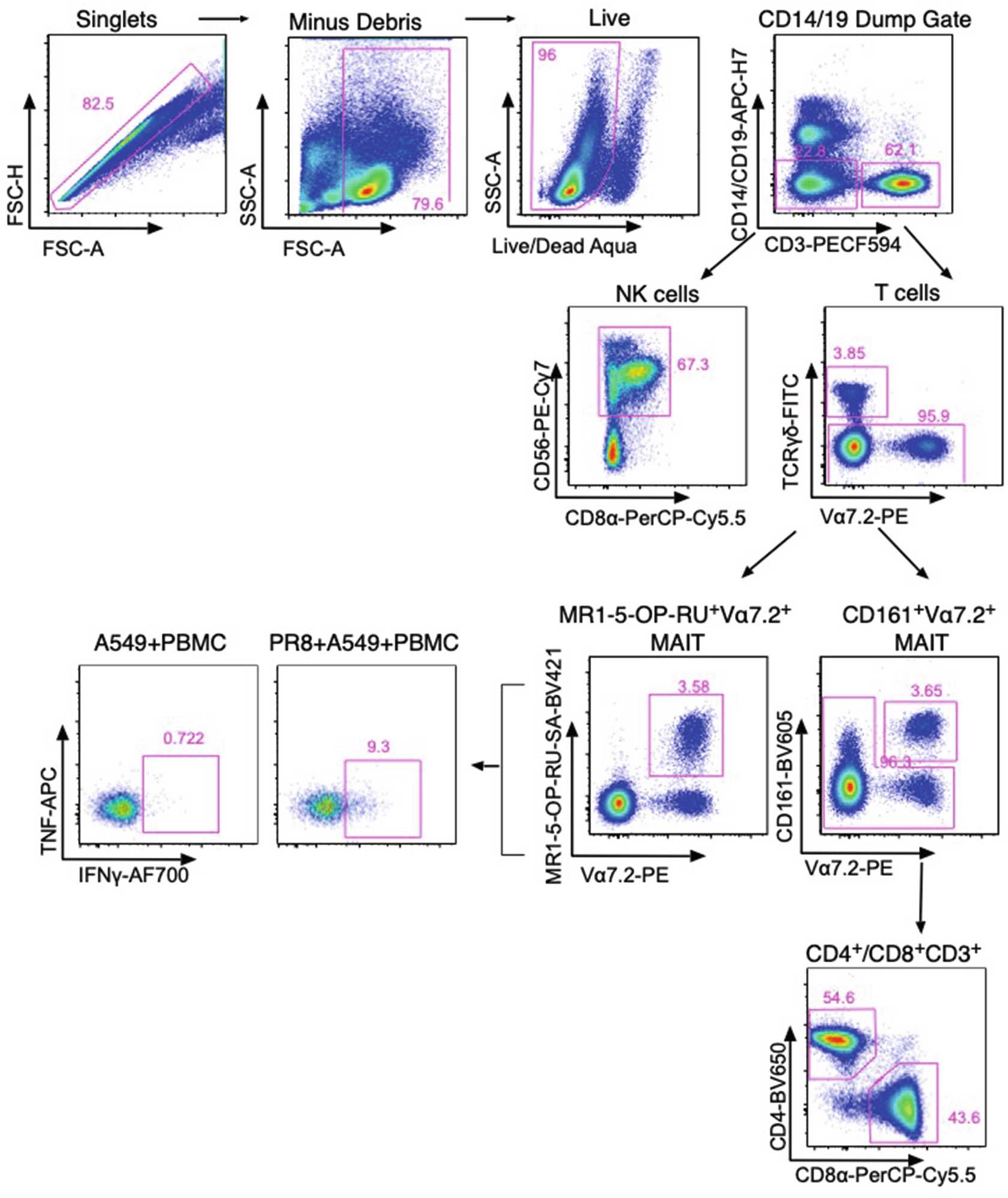
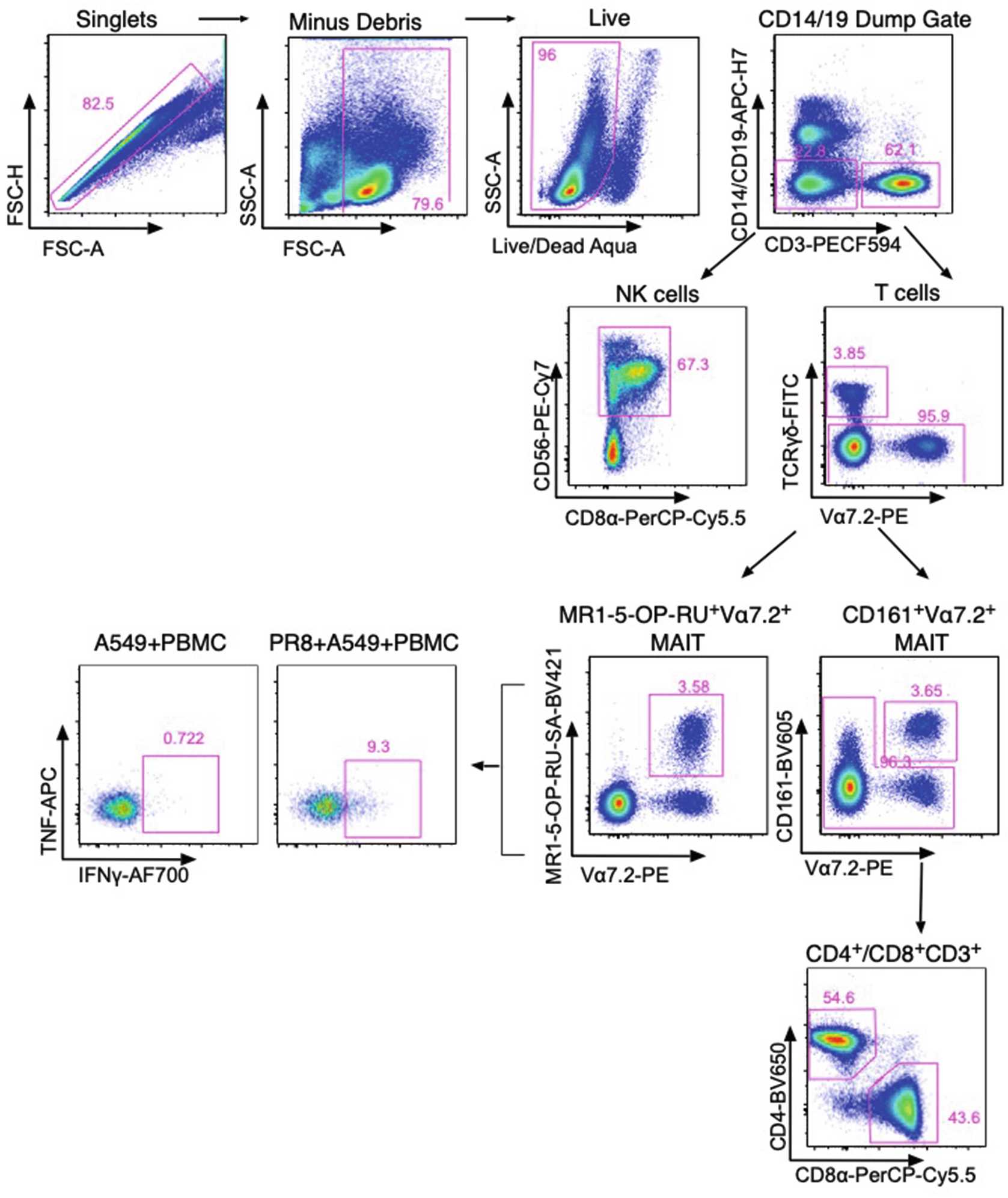
Influenza A Virus-Infected Lung Epithelial Cell Co-Culture with Human Peripheral Blood Mononuclear Cells
Liyen Loh, Marios Koutsakos, Katherine Kedzierska, Timothy S. C. Hinks
Abstract
Sensing of influenza A virus (IAV) infection by pattern recognition receptors can occur by either direct infection of lung epithelial cells or uptake of virus-infected cells by innate cells such as dendritic cells/monocytes. This triggers a series of downstream events including activation of the inflammasome, the production of cytokines, chemokines, and the upregulation of stress-induced ligands that can lead to the activation of innate cells. These cells include innate lymphocytes such as MAIT, NKT, NK, and γδ T cells. Here we describe a method used to allow activation of human innate lymphocytes in co-culture with an IAV-infected human lung epithelial cell line (A549) to measure ex vivo effector functions (TNF and IFNγ) in a mixed culture environment. We describe (1) infection of the human lung epithelial cell line, (2) co-culture with PBMC, and (3) measurement of activation using intracellular cytokine staining.
Before start
Biological Hazards—Human PBMC samples are classified as non-infectious. Influenza A virus—PR8-strain (H1N1) is a lab-adapted strain of IAV virus. Work should be risk assessed, and we recommend controls which include but are not restricted to the following: Lab coat, safety glasses, and gloves should be worn when performing this protocol. Work with human PBMCs and virus in a Class II biohazard cabinet. Use filter tips when working with virus. Decontaminate all pipette tips that have been used for human and virus work in 10% lysol or 1% Virkon when working in the biohazard cabinet. After use, the biohazard hood should be decontaminated by wiping down with 70% ethanol and by UV sterilization for 15 min before any further use. All waste and its container must be disposed as hazardous waste.
Attachments
Steps
IAV Infection of Human Lung Epithelial Cell Line, A549
24h 0m 0s prior to infection, in two T75 flasks, seed 5 x 106 A549 cells in a total volume of 20mL.
On the day of infection:
Leave one flask of A549 cells in the incubator (uninfected control).
Wash the other flask with Room temperature once, cap and gently rotate flask from side to side. Aspirate PBS with glass tissue culture pipette.
Thaw virus (PR8) On ice and add 174µL to 10mL in a 50 mL falcon tube (depending on viral titer of stock) to achieve a multiplicity of infection (MOI) of ~10–30.
Gently pipette this into the T75 containing A549 cells.
Incubate flask horizontally for 1h 0m 0s in the 37°C incubator (5% CO2).
Remove both T75 flasks from incubator and add 10mL to the flask containing virus. Cap and gently rotate from side to side. Aspirate media from both flasks.
To detach A549 cells, wash flasks once with 37Room temperature, aspirate, and add 2.5mL to each flask. Gently tilt the flask to ensure that the solution coats the entire flask.
Incubate for 0h 5m 0s in the 37°C incubator (5% CO2).
Add 10mL to T75 flasks and transfer the contents into two 50 mL falcon tubes.
Centrifuge 500x g,25°C.
Aspirate supernatant.
Resuspend cells in 2mL and perform cell counts using trypan blue estimation.
Adjust the volume of A549 cells so that the final concentration is 2x106 cells/mL.
Co-Culture (Start During the 1 h Incubation with Virus)
Thaw PBMCs in 37°C.
Gently pipette dropwise into 9mL per cryovial.
Centrifuge 500x g.
Aspirate media and count cells.
Resuspend PBMCs at 10x106 cells/mL in cRPMI.
For each sample, aliquot 100µL into three wells of a 96-well U-bottom plate.
Check IAV nucleoprotein levels:
Spin down plate by centrifuging at 400x g,4°C. Discard supernatant in waste container containing 10% Lysol or 1% Virkon in class II biosafety cabinet.
Resuspend cells in 100µL and transfer to bullet tubes.
Stain cells with live/dead discrimination marker Aqua (1:800) final volume of 50 μL/well. Use PBS as a diluent. Incubate at 4Room temperature in the dark for 0h 15m 0s.
Centrifuge plate at 400x g,4°C. Discard supernatant.
Resuspend the cells in 100µL and incubate On ice in the dark for 0h 20m 0s.
Wash cells with 100µL. Centrifuge for 450x g,4°C.
Resuspend cells in 50µL, see (Table 1) below.
Table 1 (Antibodies):
Example flow cytometric activation panel and IAV nucleoprotein expression
| A | B | C | D | E |
|---|---|---|---|---|
| Marker | Fluorophore | Laser | Clone | Dilution |
| Surface stain activation | ||||
| Live Dead | Aqua | Violet | 1/800 (stain in PBS prior to surface stain) | |
| MRI-5-OP-RU Tet | SA-BV421 | Violet | *Titrate 1/200-1/400 | |
| CD19 | APC-H7 | Red | H1B19 | 1/100 |
| CD14 | APC-H7 | Red | MΦP9 | 1/100 |
| CD8α | PerCP-Cy5.5 | Blue | SK1 | 1/50 |
| TCRγδ | FITC | Blue | 2F11 | 1/50 |
| CD4 | BV650 | Violet | OKT4 | 1/200 |
| CD161 | BV605 | Violet | HP-3G10 | 1/50 |
| CD3 | PE-CF594 | Yellow/Green | UCHT1 | 1/200 |
| TCR Vα7.2 | PE | Yellow/Green | 3C10 | 1/200 |
| CD56 | PE-Cy7 | Yellow/Green | NCAM16.2 | 1/100 |
| Intracellular stain activation | ||||
| TNF | APC | Red | MAb11 | 1/50 |
| IFNγ | AF700 | Red | B27 | 1/100 |
| Intracellular stain IAV-NP infection | ||||
| IAV nucleoprotein | FITC | Blue | 1331 | 1/100 |
Typical flow cytometry panel compatible with a four-laser BD LSRII Fortessa flow cytometer, allowing identification of innate and adaptive lymphocyte subsets and assessment of activation measured by intracellular cytokine staining *Batches of SA-conjugated and Tetramerized MR1-5-OP-RU will vary and require titration prior to usage
Incubate On ice in the dark for 0h 30m 0s.
Wash cells with 150µL. Centrifuge for 450x g,4°C.
Repeat with a second wash with 200µL. Centrifuge for 450x g,4°C.
Add 100µL of infected and uninfected A549 cells to separate wells in the 96-well plate.
Add 100µL of uninfected A549s or IAV-infected A549s (2x105 cells) into wells containing PBMC. Leave one well with PBMC only, add 100µL to this well. Place plate in the 37°C incubator (5% CO2).
After 3h 0m 0s–4h 0m 0s, add brefeldin A (BFA-GOLGI PLUG), 1:2000 to all wells.
Incubate for a further 6h 0m 0s in the 37°C incubator (10h 0m 0s).
Remove plate and continue with intracellular cytokine (ICS) staining or place in the 4°C covered in foil to stain the next day.
Intracellular Cytokine Staining
Spin down plate by centrifuging at 400x g,4°C. Discard supernatant in waste container containing 10% Lysol or 1% Virkon in class II biosafety cabinet.
Stain cells with live/dead discrimination marker Aqua (1:800) final volume of 50 μL/well. Use PBS as a diluent. Incubate at 4Room temperature in the dark for 0h 15m 0s.
Centrifuge plate at 400x g,4°C. Discard supernatant.
Add 50µL (Table 1 below) to each well.
Table 1 (Antibodies):
Example flow cytometric activation panel and IAV nucleoprotein expression
| A | B | C | D | E |
|---|---|---|---|---|
| Marker | Fluorophore | Laser | Clone | Dilution |
| Surface stain activation | ||||
| Live Dead | Aqua | Violet | 1/800 (stain in PBS prior to surface stain) | |
| MRI-5-OP-RU Tet | SA-BV421 | Violet | *Titrate 1/200-1/400 | |
| CD19 | APC-H7 | Red | H1B19 | 1/100 |
| CD14 | APC-H7 | Red | MΦP9 | 1/100 |
| CD8α | PerCP-Cy5.5 | Blue | SK1 | 1/50 |
| TCRγδ | FITC | Blue | 2F11 | 1/50 |
| CD4 | BV650 | Violet | OKT4 | 1/200 |
| CD161 | BV605 | Violet | HP-3G10 | 1/50 |
| CD3 | PE-CF594 | Yellow/Green | UCHT1 | 1/200 |
| TCR Vα7.2 | PE | Yellow/Green | 3C10 | 1/200 |
| CD56 | PE-Cy7 | Yellow/Green | NCAM16.2 | 1/100 |
| Intracellular stain activation | ||||
| TNF | APC | Red | MAb11 | 1/50 |
| IFNγ | AF700 | Red | B27 | 1/150 |
| Intracellular stain IAV-NP infection | ||||
| IAV nucleoprotein | FITC | Blue | 1331 | 1/100 |
Typical flow cytometry panel compatible with a four-laser BD LSRII Fortessa flow cytometer, allowing identification of innate and adaptive lymphocyte subsets and assessment of activation measured by intracellular cytokine staining *Batches of SA-conjugated and Tetramerized MR1-5-OP-RU will vary and require titration prior to usage
Incubate for 0h 30m 0s On ice, in the dark.
Wash cells once with 150µL. Centrifuge for 1500rpm,4°C. Flick off supernatant in discard container in biohazard cabinet.
Resuspend the cells in 100µL and incubate On ice in the dark for 0h 20m 0s.
Wash cells with 100µL. Centrifuge for 450x g,4°C.
Resuspend cells in 50µL, see (Table 1) below.
Table 1 (Antibodies):
Example flow cytometric activation panel and IAV nucleoprotein expression
| A | B | C | D | E |
|---|---|---|---|---|
| Marker | Fluorophore | Laser | Clone | Dilution |
| Surface stain activation | ||||
| Live Dead | Aqua | Violet | 1/800 (stain in PBS prior to surface stain) | |
| MRI-5-OP-RU Tet | SA-BV421 | Violet | *Titrate 1/200-1/400 | |
| CD19 | APC-H7 | Red | H1B19 | 1/100 |
| CD14 | APC-H7 | Red | MΦP9 | 1/100 |
| CD8α | PerCP-Cy5.5 | Blue | SK1 | 1/50 |
| TCRγδ | FITC | Blue | 2F11 | 1/50 |
| CD4 | BV650 | Violet | OKT4 | 1/200 |
| CD161 | BV605 | Violet | HP-3G10 | 1/50 |
| CD3 | PE-CF594 | Yellow/Green | UCHT1 | 1/200 |
| TCR Vα7.2 | PE | Yellow/Green | 3C10 | 1/200 |
| CD56 | PE-Cy7 | Yellow/Green | NCAM16.2 | 1/100 |
| Intracellular stain activation | ||||
| TNF | APC | Red | MAb11 | 1/50 |
| IFNγ | AF700 | Red | B27 | 1/150 |
| Intracellular stain IAV-NP infection | ||||
| IAV nucleoprotein | FITC | Blue | 1331 | 1/100 |
Typical flow cytometry panel compatible with a four-laser BD LSRII Fortessa flow cytometer, allowing identification of innate and adaptive lymphocyte subsets and assessment of activation measured by intracellular cytokine staining *Batches of SA-conjugated and Tetramerized MR1-5-OP-RU will vary and require titration prior to usage
Incubate On ice in the dark for 0h 30m 0s.
Wash cells with 150µL. Centrifuge for 450x g,4°C.
Repeat with a second wash with 200µL. Centrifuge for 450x g,4°C.
Resuspend cells in 100µL and transfer to bullet tubes.