In Silico analysis links the NSL complex to Parkinson’s disease and the mitochondria – Protein-protein interaction data to functional enrichment analysis
Katie Kelly, c.manzoni, Patrick Lewis, Helene Plun-Favreau
Parkinson’s disease
NSL complex
Mitophagy
In silico
Protein-protein interaction (PPI)
Mito-CORE network Interactome
ASAPCRN
Abstract
Whilst the majority (~90-95%) of PD cases are sporadic, much of our understanding of the pathophysiological basis of disease can be traced back to the study of rare, monogenic forms of disease. However, in the past decade, the availability of Genome-Wide Association Studies (GWAS) has facilitated a shift in focus, toward identifying common risk variants conferring an increased risk of developing PD across the population.
A recently developed mitophagy screening assay of GWAS candidates, has functionally implicated the non-specific lethal (NSL) complex, a chromatin remodeler, in the regulation of PINK1-mitophagy. Here, a bioinformatics approach has been taken to investigate the interactome of the NSL complex, to unpick its relevance to PD progression. The mitochondrial interactome of the NSL complex has been built, mining 3 separate repositories: PINOT, HIPPIE and MIST, for curated, literature-derived protein-protein interaction (PPI) data. A multi-layered approach has been taken to; i) build the ‘mitochondrial’ NSL interactome, applying PD gene-set enrichment analysis to explore the relevance of the NSL mitochondrial interactome to PD and, ii) build the PD-oriented NSL interactome, using functional enrichment, to uncover biological pathways underpinning the NSL /PD association.
Steps
Downloading and merging the Protein-Protein Interaction (PPI) Data
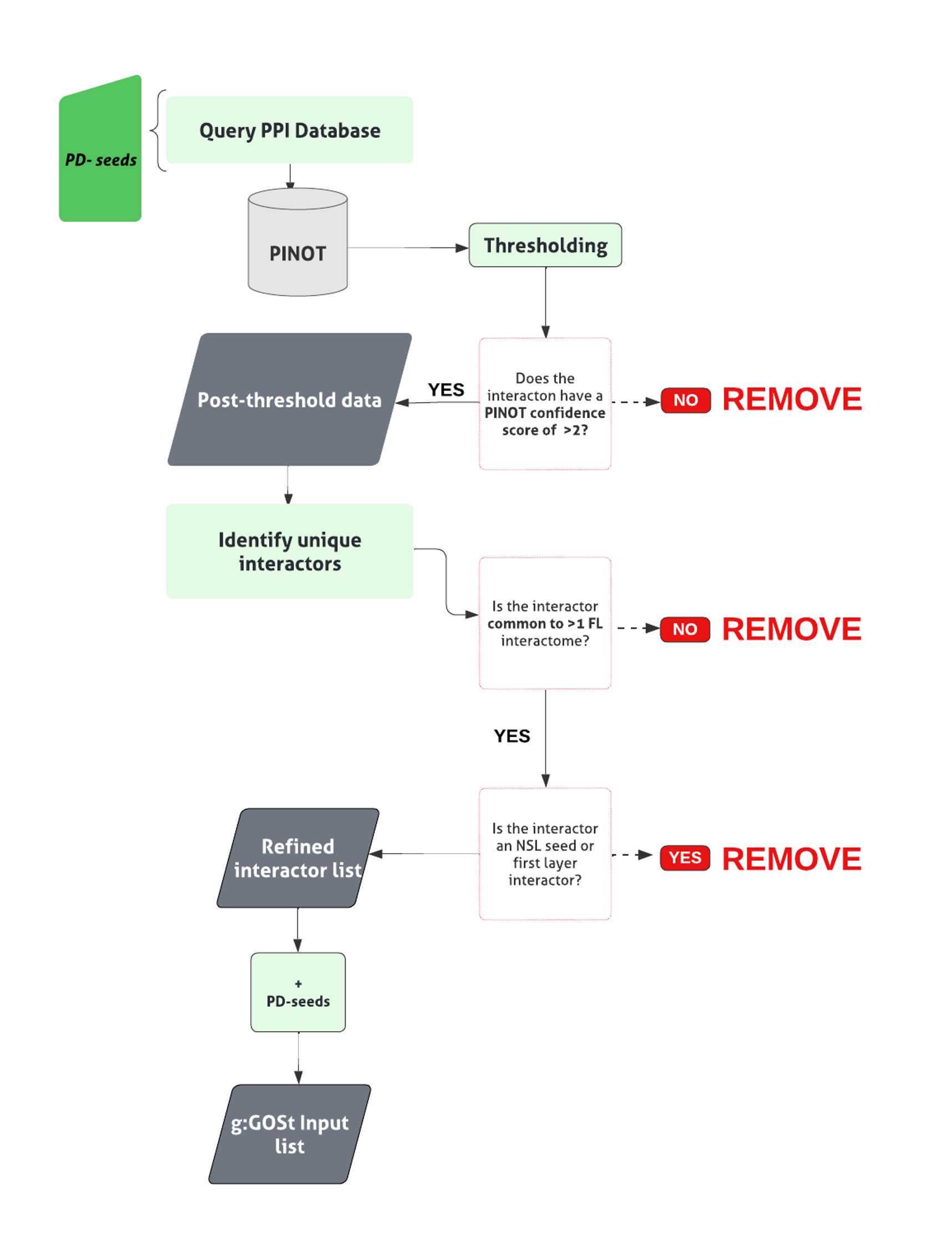
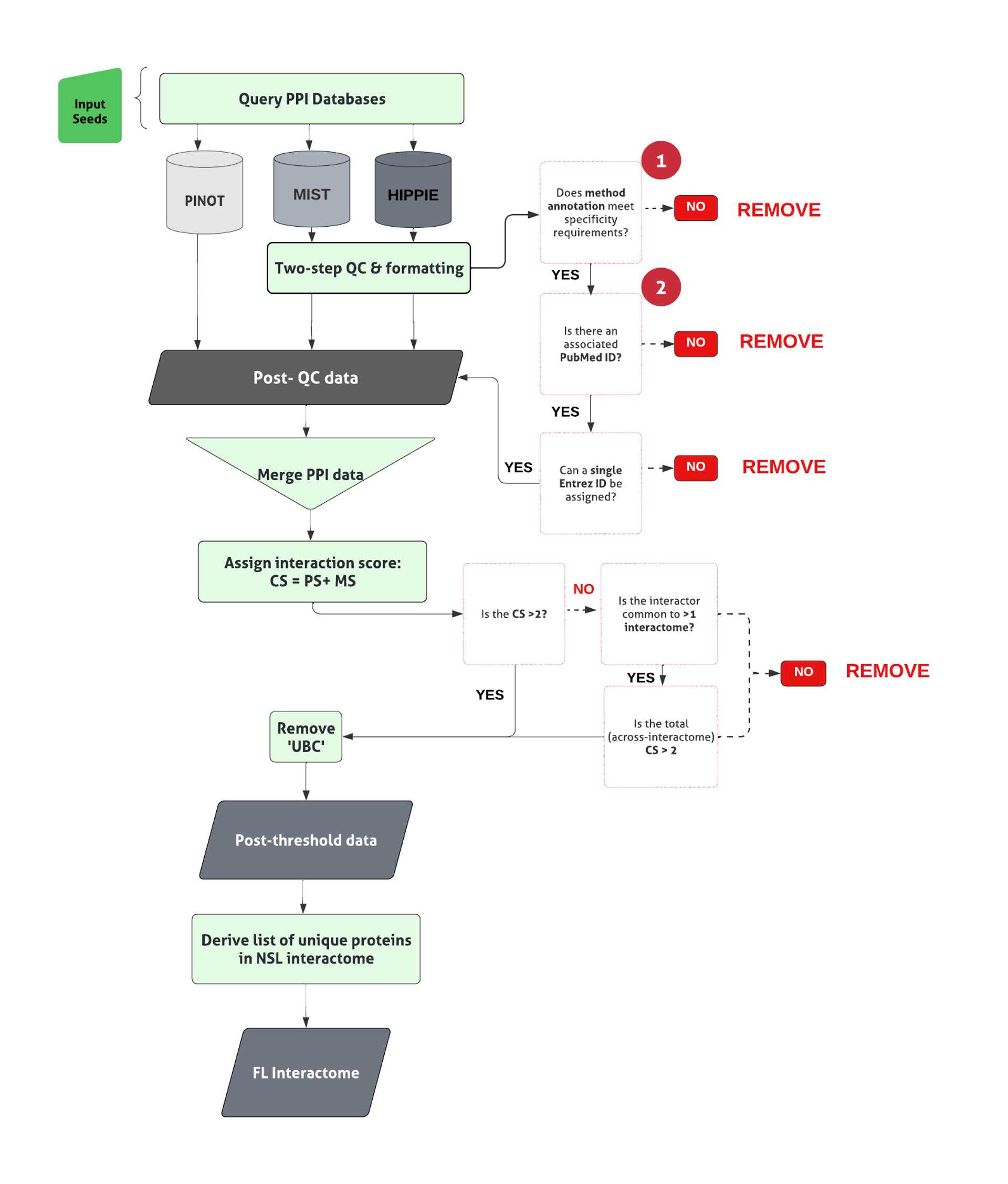
All code can be found here : 10.5281/zenodo.7875446.The general pipeline to derive the first layer first layer interactome can be found in Figure 1.
Collect PPIs for NSL seeds using 3 different web-based tools;
-
PINOT (Version 1.1 with lenient filter option) (Protein Interaction Network Online Tool) (Tomkins, Ferrari et al. 2020, DOI: http://dx.doi.org/10.1186/s12964-020-00554-5)
-
HIPPIE with no threshold on interaction score (Human Integrated Protein-Protein Interaction rEference) (Alanis-Lobato, Andrade-Navarro et al. 2017 ; DOI: https://doi.org/10.1093/nar/gkw985; RRID:SCR_014651).
-
MIST v5.0 (Molecular Interaction Search Tool) (Hu, Vinayagam et al. 2018 ; DOI: 10.1093/nar/gkx1116).
PPI data obtained using MIST and HIPPIE are subjected to quality control (QC), QC steps 1 & 2 (already integrated within the PINOT pipeline) to remove low quality data.
Formatting between the output files is standardized and interactors’ IDs are converted to the approved EntrezID, UniprotID and HGNC gene name.
Prior to merging the results for each interaction, files are parsed to identify the number of times the interaction was i) observed via a unique methodological technique and ii) reported in a unique publication.
Apply PINOT method grouping to the interactions downloaded from HIPPIE and MIST, to ensure consistency between the results from each database. To do so, download the 'Method conversion table' ; https://www.reading.ac.uk/bioinf/PINOT/PINOT_help.html#select from PINOT and convert methods according to the MI code.
Parse files from each database to generate a separate dataframe containing 'publication' observations (for calculation of the publication score (PS)), and 'method' observations (for calculation of the method score (MS)). Unique observations for each interaction in each dataframe are allocated an individual row.
Thresholding the PPIs
Merge 'Publication' observations and 'method' observation files. The number of rows occupied by each interaction corresponds to the number of observations. The CS for each interaction can be calculated calculated as:
Apply a score threshold ( CS >2), to filter and remove lower confidence PPI data lacking reproducibility.
If interactions that failed to meet the threshold, interrogate further, to identify those interactors bridging >1 interactome.
For those interactors appearing within >1 interactome, apply a multi-interactome threshold represented by a CS > 2 across interactomes. Retain those meeting this multi-interactome threshold.
Combine those interactions meeting the single and multi- interactome threshold, to generate the first layer interactome.
Where ‘UBC’, a ubiquitin moiety, is identified as an interactor within the first layer , review the supporting publication. Unless the interaction being studied is specific, remove.
Generate the list of unique interactors within the first layer interactome
Generating the Mito-CORE Network
The pipeline to derive the Mito-CORE network can be found in Figure 2.
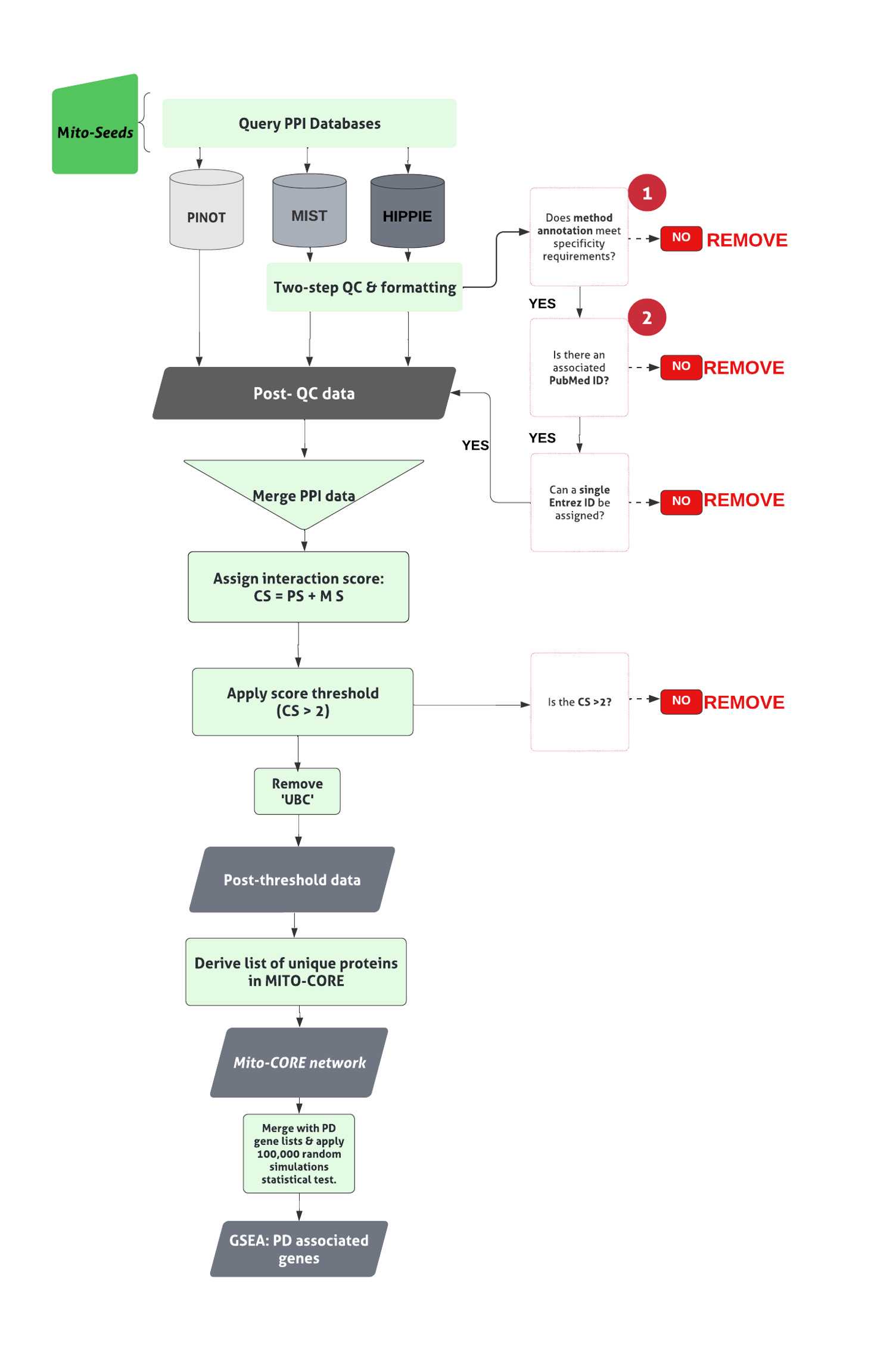
First, prioritise members of the first layer with mitochondrial annotation (- OGT, since it was a seed to derive the first layer interactome). Here, these have been termed ‘ Mito seeds’.
Merge each list of mitochondrial proteins with the first layer interactome, to find overlaps. The overlaps represent members of the mitochondrial interactome for the NSL complex.
Input mito seeds into all three PPI tools, to obtain the second layer . The NSL seeds together with the Mito seeds , and second layer interactors form the complete Mito-CORE network.
Gene Set Enrichment Analysis (GSEA)
Conduct GSEA for PD associated genes by comparing the members of the interactome under investigation ( first layer alone or complete Mito-CORE network) to a list of 180 unique PD associated genes;
Merge the list of 180 PD associated genes with the list of unique ( first layer / Mito-CORE network) interactors, to find overlaps between the two lists. The overlaps represent PD associated proteins within the direct interactome/mitochondrial interactome for the NSL complex
Repeat the above step with the list of 15 Mendelian PD genes, to ascertain enrichment of this more stringent list.
Statistical Evaluation via Random Networks Simulation
Use an ‘100,000 random simulations’ test of significance to validate statistical significance of overlaps of PD genes with the first layer and complete Mito-CORE network (code found in file 100,000 Random Simulations testing (GitHub) ).
Generating the PD-CORE Network
Input PD seeds into PINOT to obtain the second layer of the PD-CORE network.
Apply an arbitrary confidence threshold ' CS >2', eliminating data with just a single publication and method from the downstream analysis
Once again, convert interactors’ IDs to the approved EntrezID, UniprotID and HGNC gene name.
To remove background noise, keep only members of the second layer bridging >1 PD seed within the PD-CORE network.
The NSL seeds together with the PD seeds, and the non-private second layer interactors from the complete PD-CORE network .
Functional Enrichment Analysis
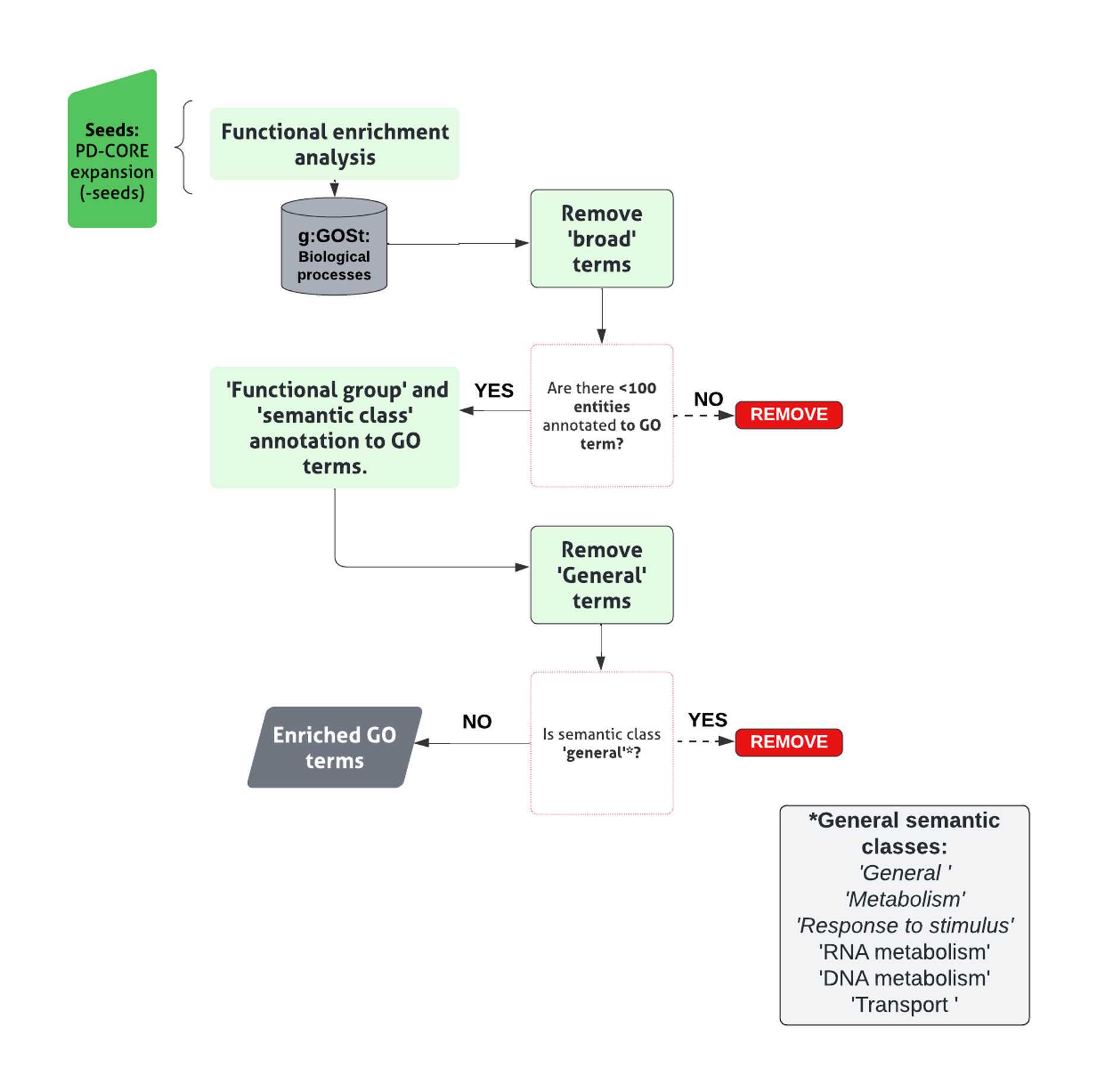
Assess enrichment of particular biological processes within the PD-CORE network, members (- NSL seeds), by inputting into the g:Profiler search tool, g:GOSt (G:Profiler ; Ashburner, Ball et al. 2000, Gene Ontology 2021; RRID:SCR_006809).
Conduct enrichment for GO terms associated with ‘Biological Processes (BPs)’ only, with all other analysis settings left unadjusted, generating a list of enriched GO:BP terms.
Apply a threshold to the list of enriched GO:BP terms, to retain those with term size <100 thus effectively removing ‘broad’ GO:BP terms.
Assign remaining terms to custom-made ‘semantic classes’(SC), accompanied by a parent ‘functional group’(FG) and discard generic terms.
Pool GO:BP terms contributing to each semantic class to identify the list of proteins within the network contributing to the enrichment of that specific semantic class.
The final list of semantic classes, within each functional group represents those enriched within the network.