In Silico Molecular Docking with Ligand Target
Angelo José Rinaldi
Molecular Docking
Enzyme-Ligand Interaction
Protein Data Bank
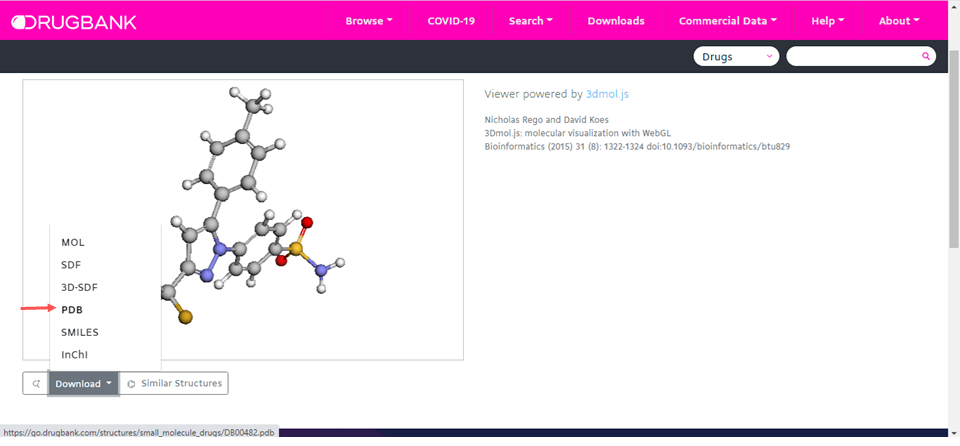
DrugBank
UCSF Chimera
MetaPocket 2.0
AutoDockTools
AutoDock Vina
ProteinsPlus
Bioinformatics
Drug Discovery.
Cyclooxygenase
Celecoxib
Abstract
This study outlines a comprehensive molecular docking protocol aimed at exploring enzyme-ligand interactions, leveraging various bioinformatics tools and databases. The protocol commences with the acquisition of the enzyme's three-dimensional structure from the Protein Data Bank (PDB) and the identification of potential ligands from the DrugBank database. Structural refinement and preparation of the enzyme and ligands are conducted using UCSF Chimera, ensuring proper protonation states and the addition of missing atoms. MetaPocket 2.0 is employed to predict and identify potential binding sites on the enzyme, facilitating the focus of the docking simulations on relevant regions.
The prepared structures are then processed using AutoDockTools to generate the necessary input files, including the grid parameter file (GPF) and docking parameter file (DPF). AutoDock Vina is subsequently used to perform the docking simulations, predicting the binding poses and affinities of the ligands within the identified binding sites. The docking results are analyzed and visualized using the ProteinsPlus platform, providing detailed insights into the interaction profiles and enabling the identification of key residues involved in binding.
By integrating these diverse computational tools and databases, the protocol offers a robust framework for studying enzyme-ligand interactions, which is crucial for drug discovery and the understanding of biochemical pathways. The streamlined approach enhances the accuracy and efficiency of docking studies, facilitating the identification of promising therapeutic candidates.
Steps
- In this protocol we will use as an example of molecular docking, a target protein (receptor) called cyclooxygenase, which is an important enzyme in inflammatory processes. And as a ligand, the anti-inflammatory celecoxib.
Target receptor search:
- Seeking the three-dimensional (3D) structure of the receiver from experimental data or computational models, here we will use the PDB.
- We search the PDB for the enzyme called cyclooxygenase and choose the one with the best resolution, which in this case was 5F19.

- Go in 3D view.

- On the right tab, under display files, extract the receiver in pdb format and save it in a working folder.

Target ligand search:
Preparation of the receptor:
-
Download the Chimera program from the link: https://www.cgl.ucsf.edu/chimera/download.html
-
Open the figure saved on the computer in Chimera or use the fetch id of 5F19

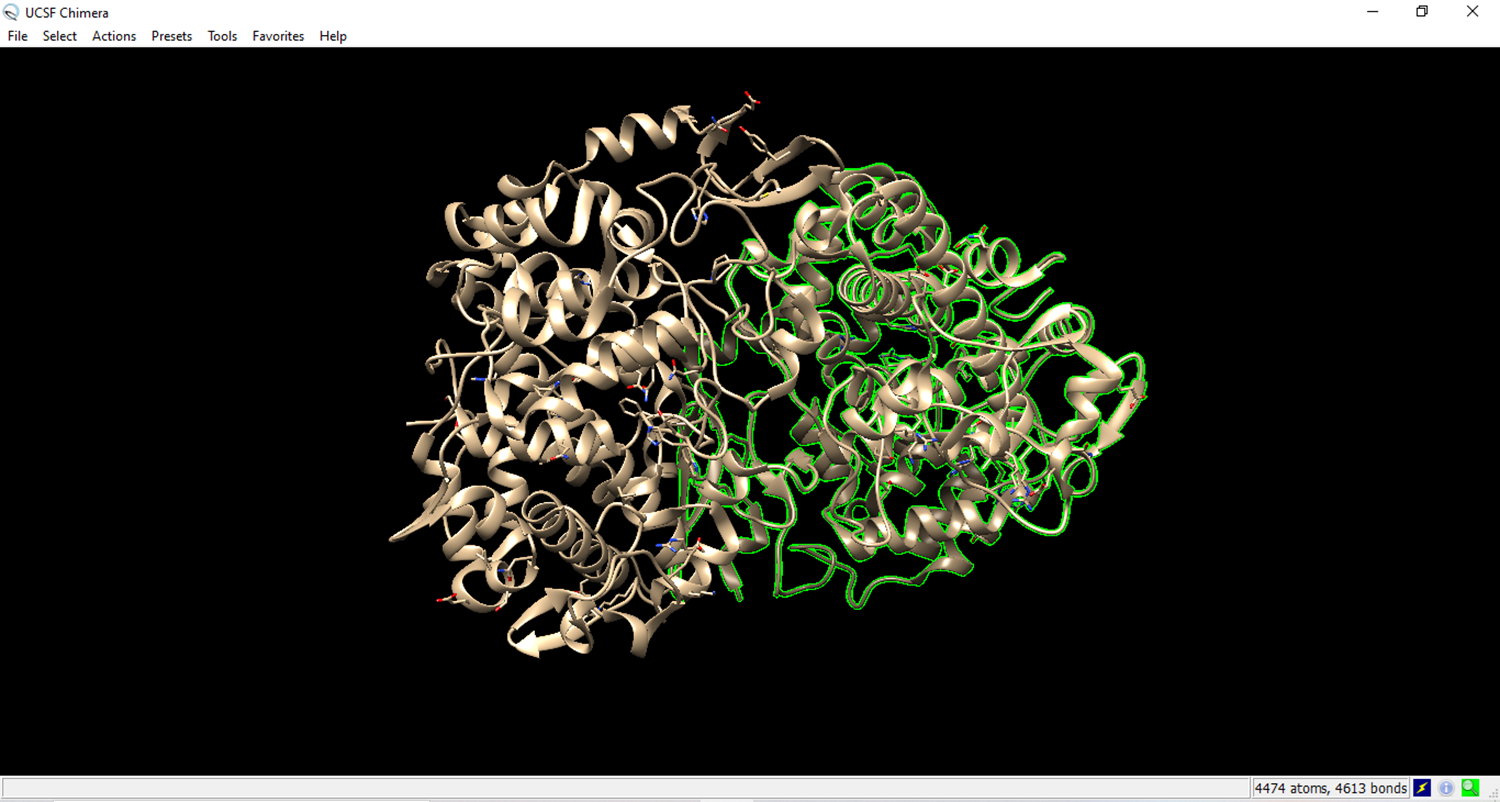
- Visualization of the cyclooxygenase enzyme (receptor) in Chimera.
- Go to select, go to residue and set to all non standard.
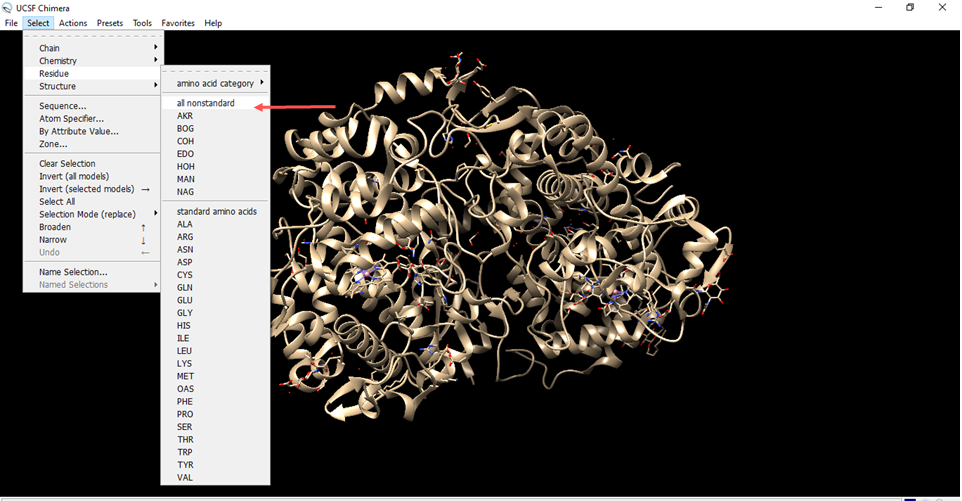
- Residues that are not from the receiver were marked in green.
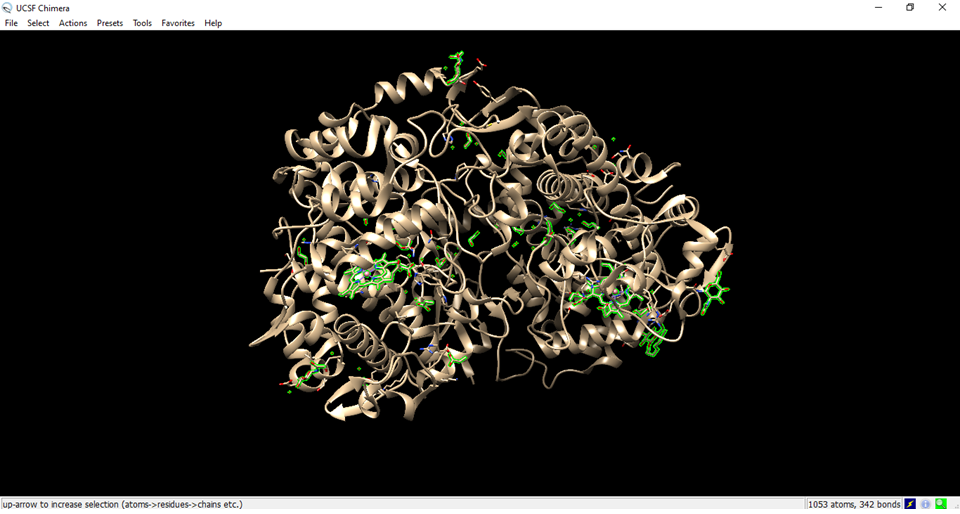
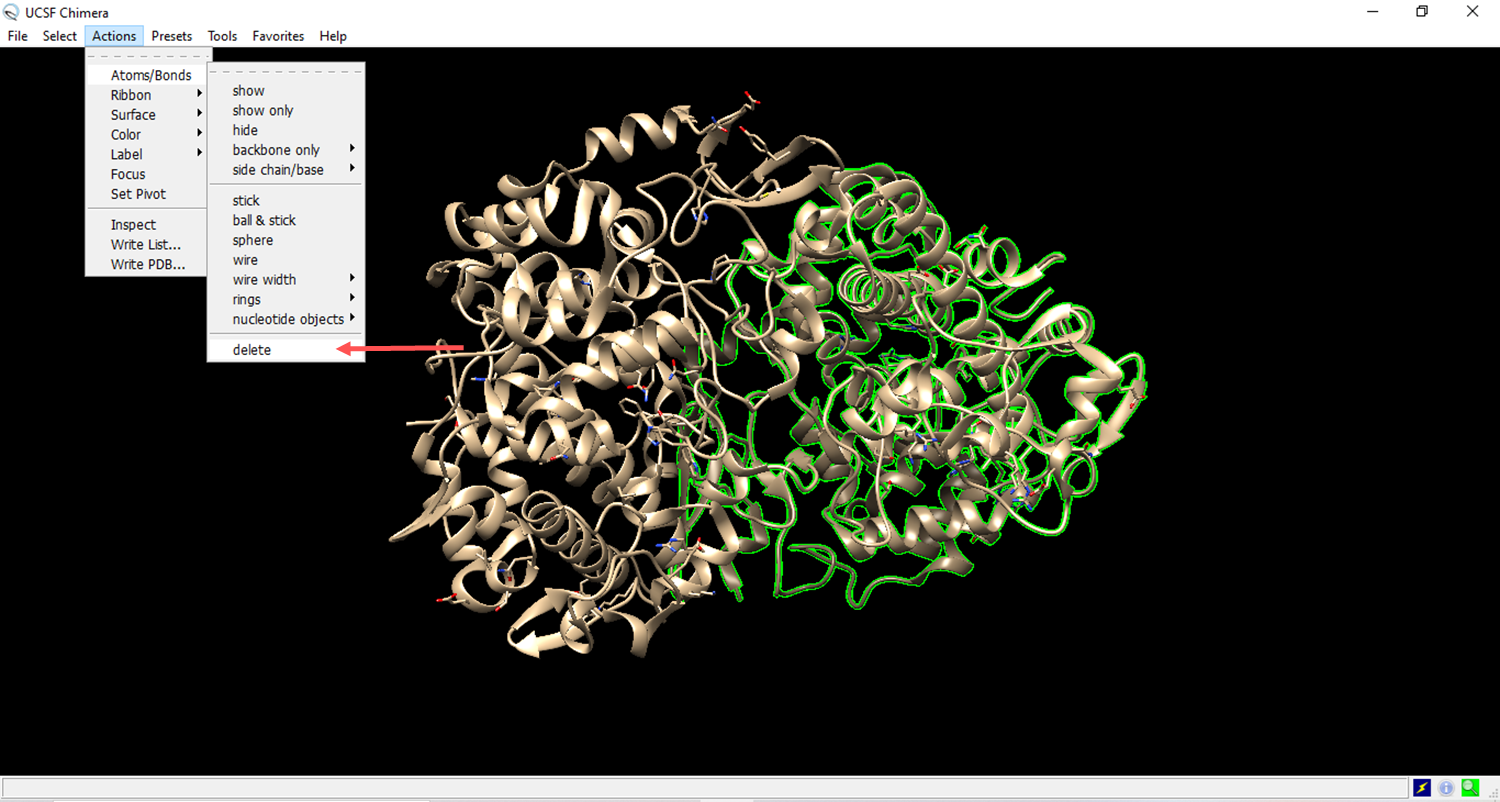
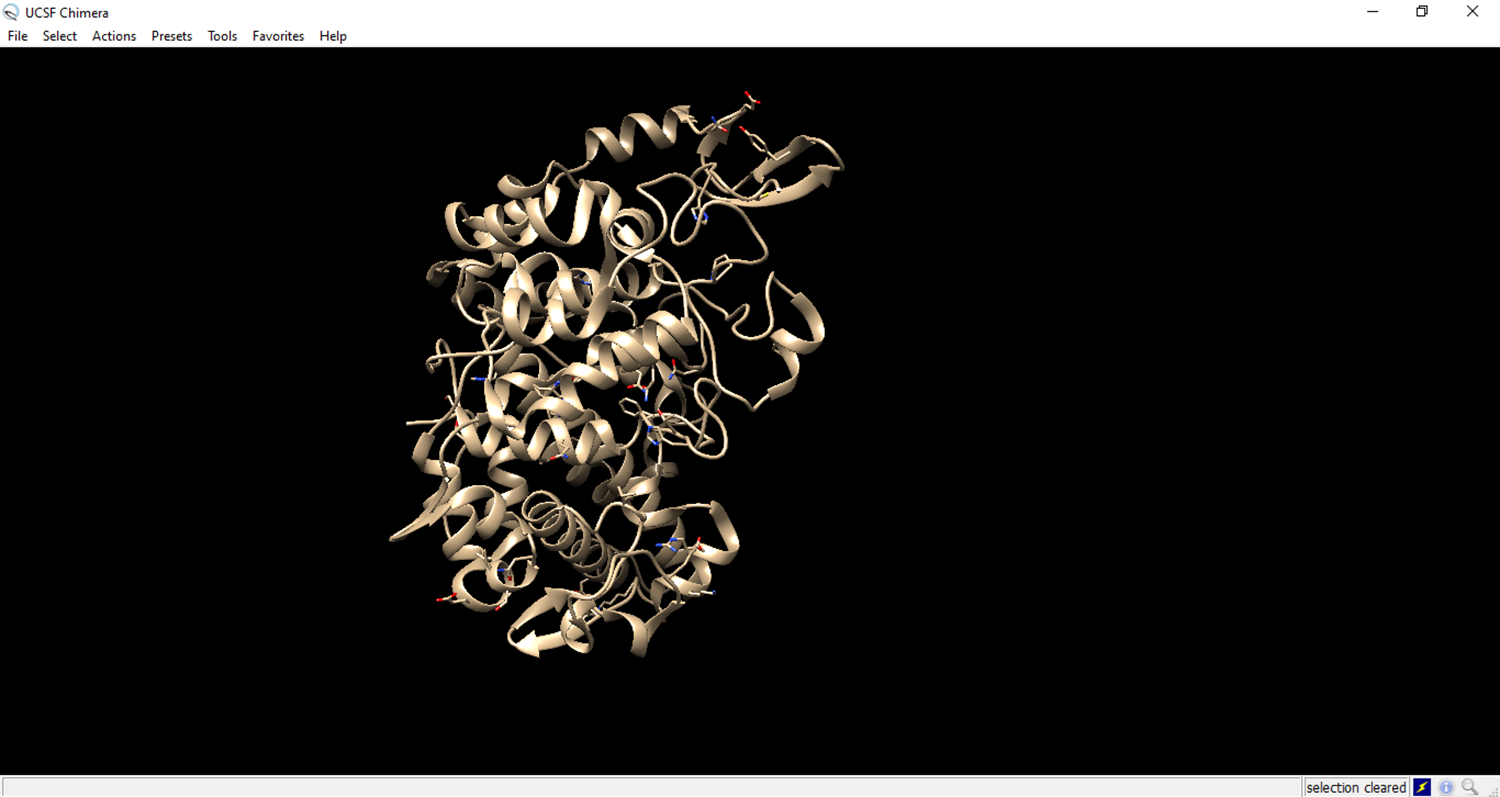
- Clean the structure
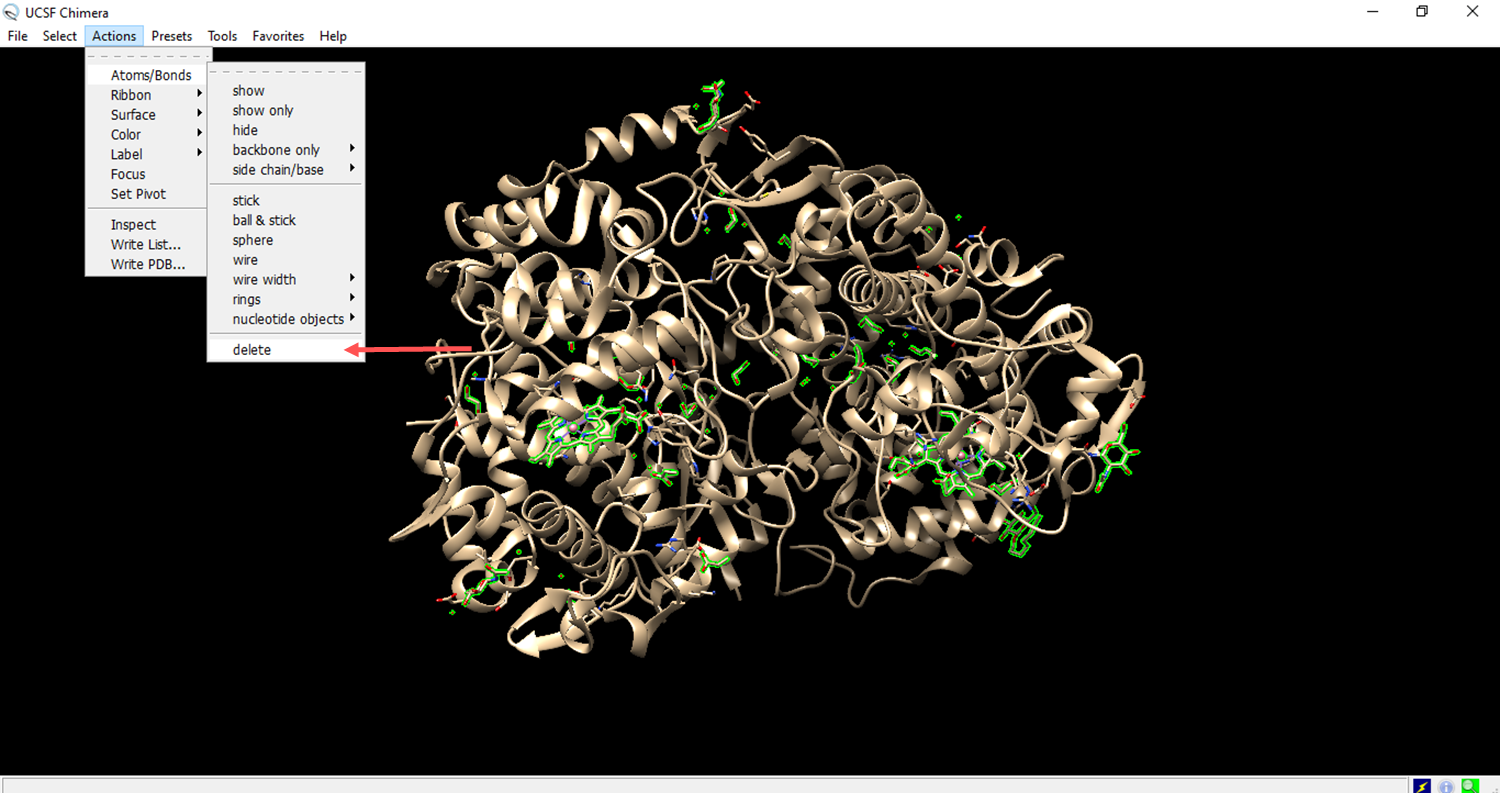
- Deleting a chain of amino acids to expose the ligand receptor.
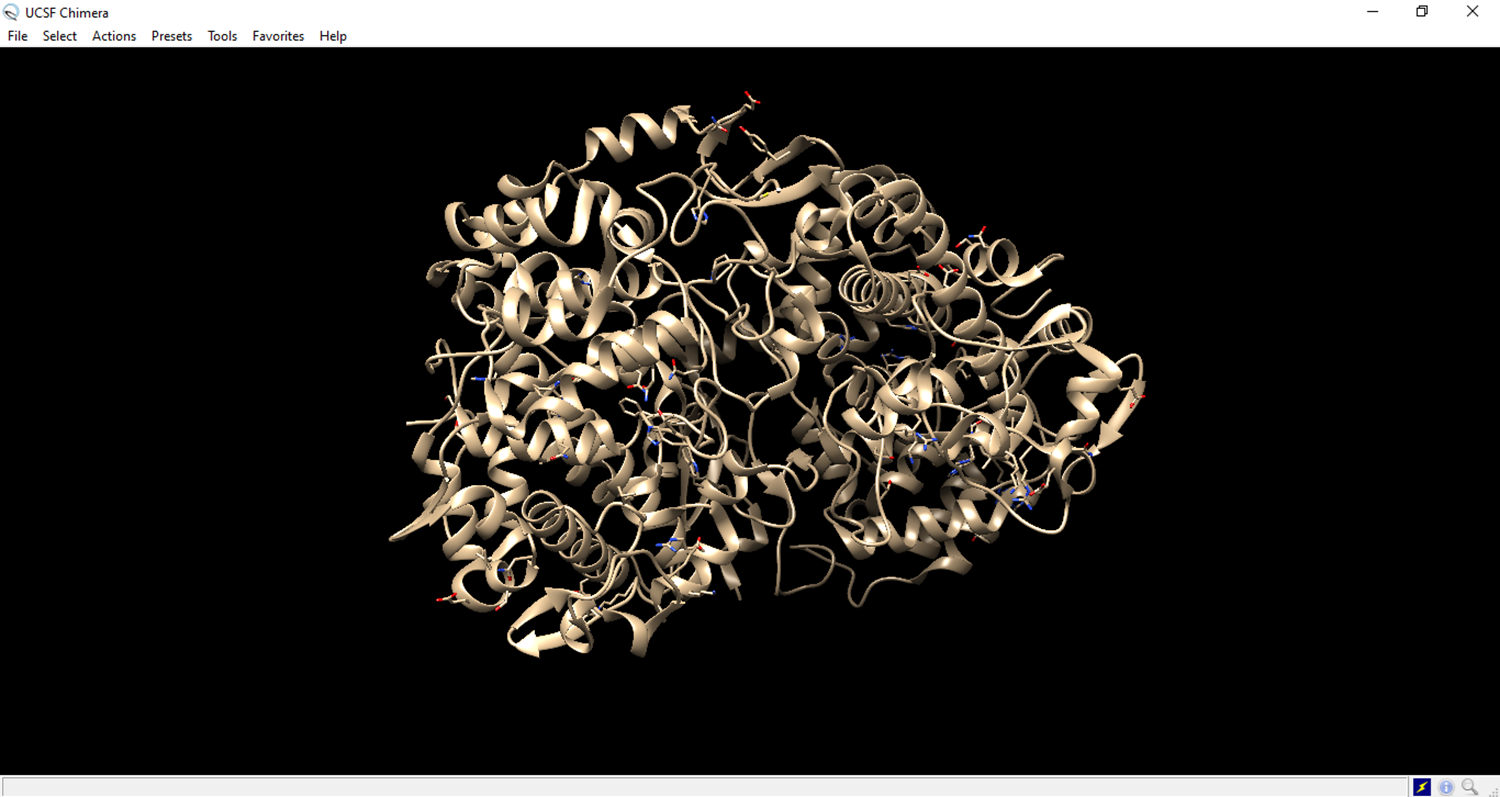
- The residues to be deleted are marked in green.
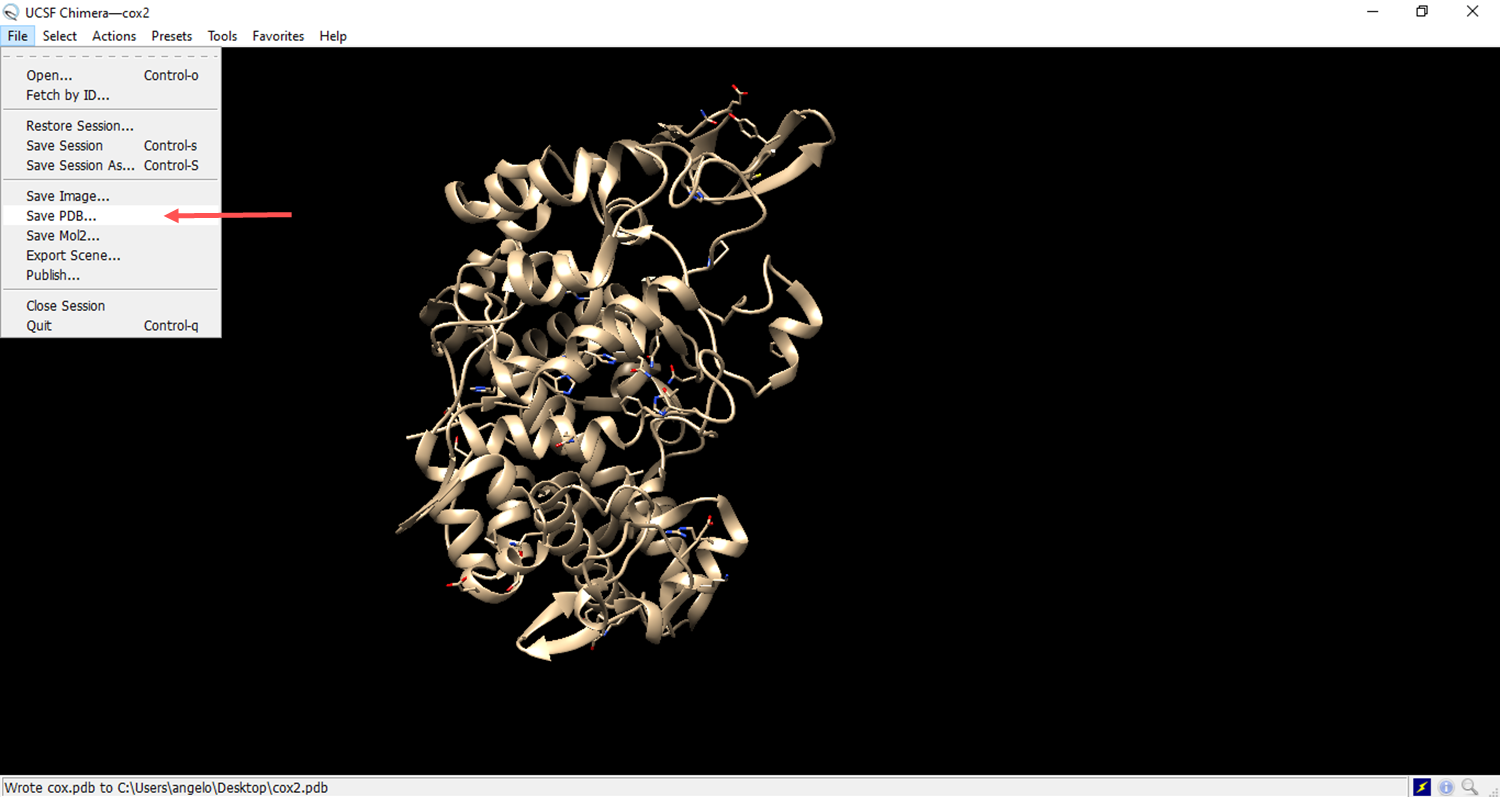
- Save the file in pdb format.
- MetaPocket 2.0 is an online tool used to predict binding sites on proteins. https://metapocket.eml.org/
Provide the structure of the prepared target protein in PDB (Protein Data Bank) format.

- Select item 4 to choose the best results
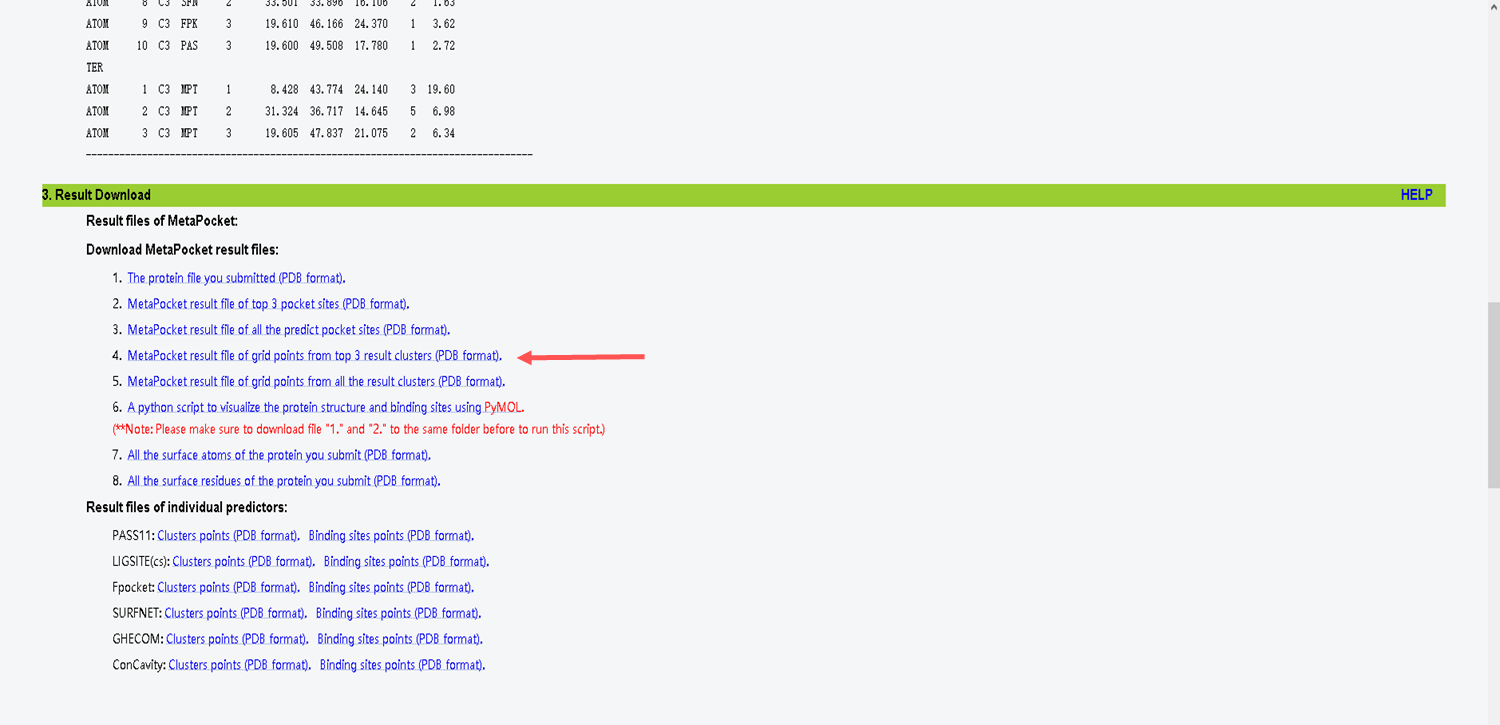
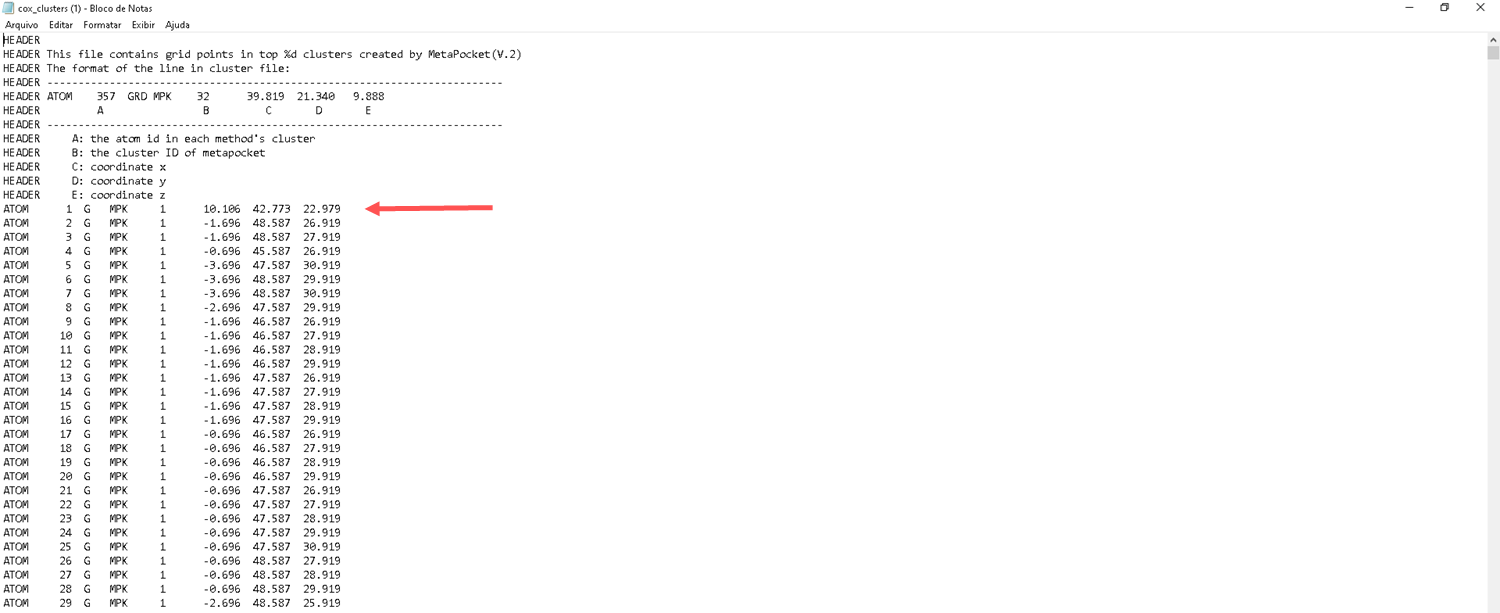
- Choose the first one that has the best result
- Tools to prepare structures, such as AutoDockTools
Download: https://autodocksuite.scripps.edu/adt/
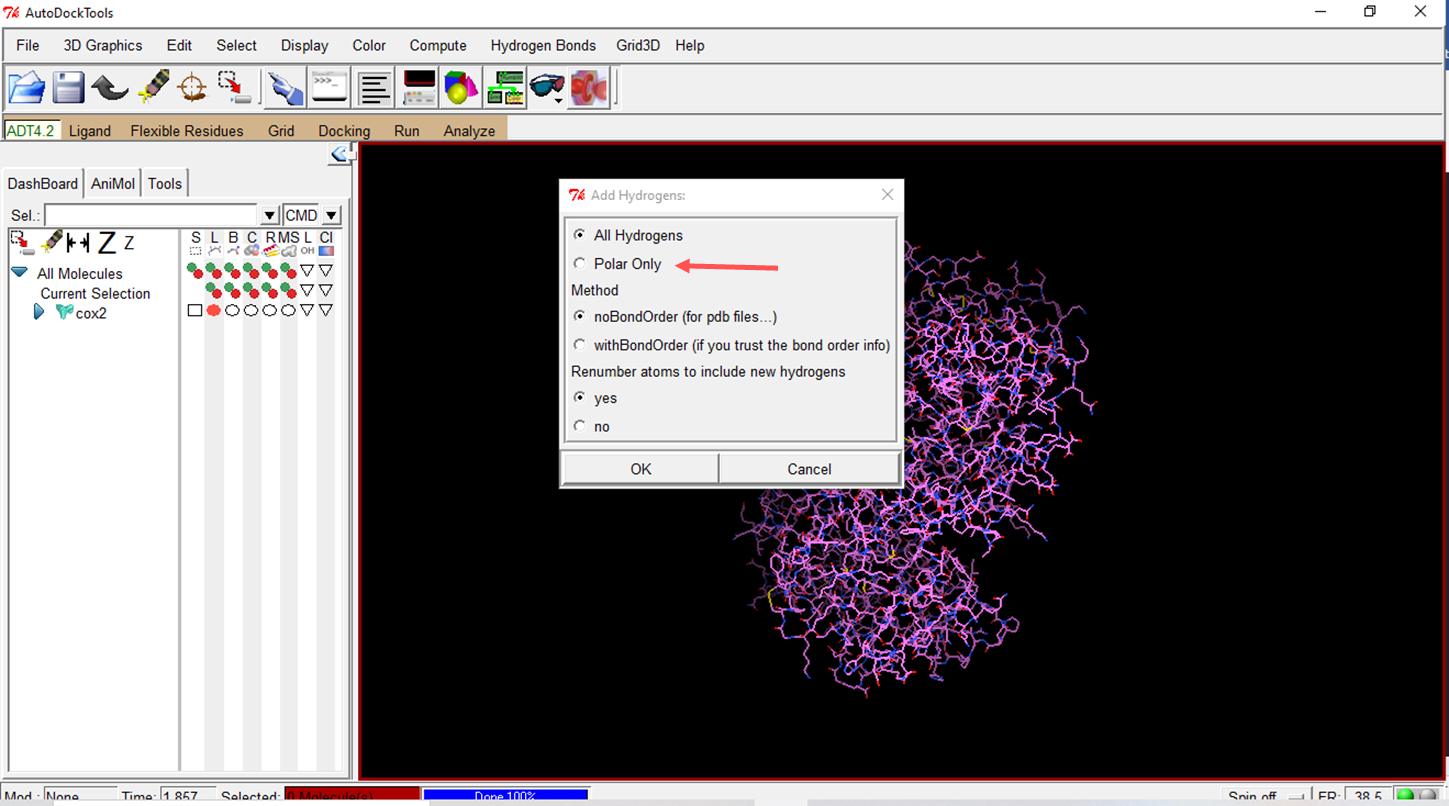
- Add hydrogens to the molecule
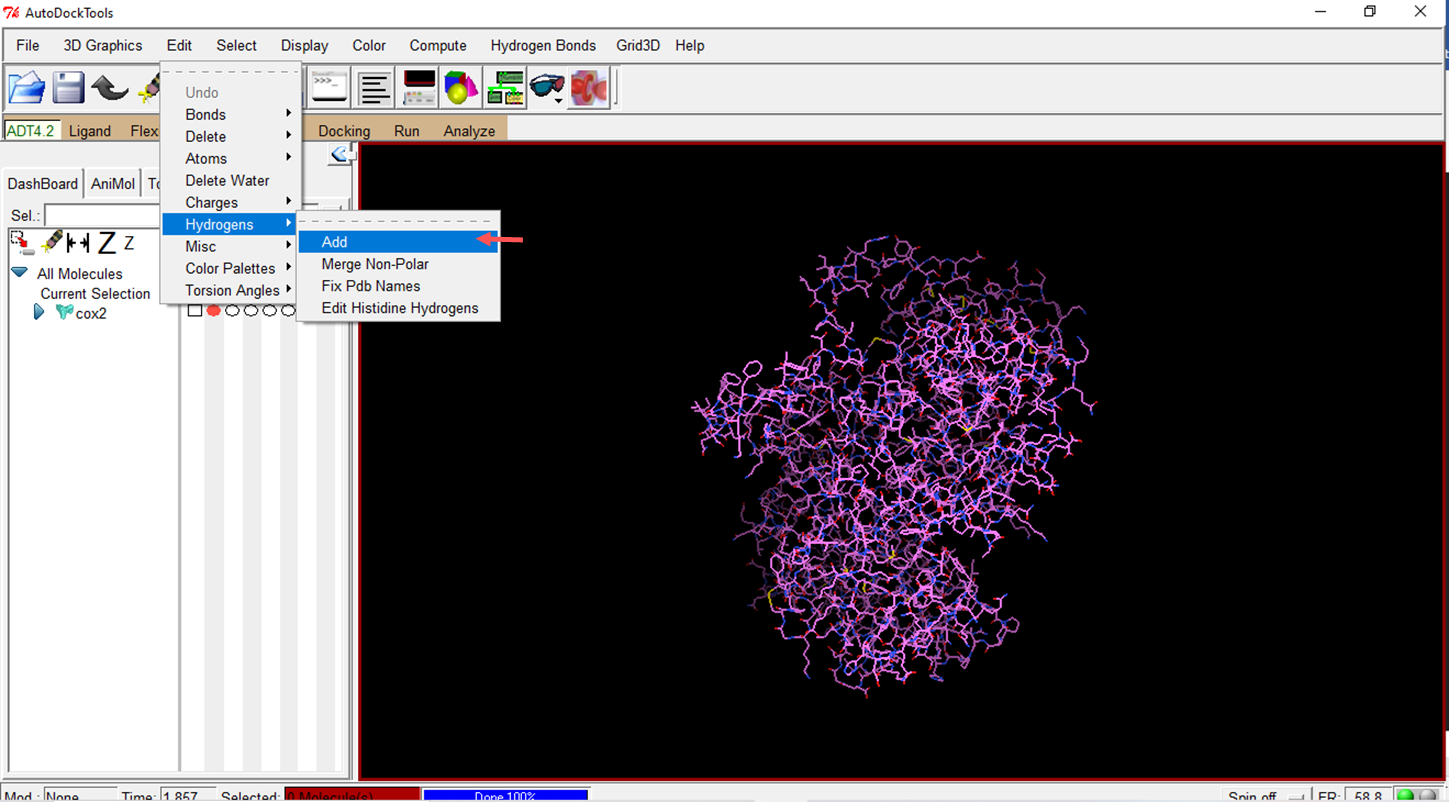
- Using the "polar only" option means that only polar hydrogens are considered when preparing the molecules.
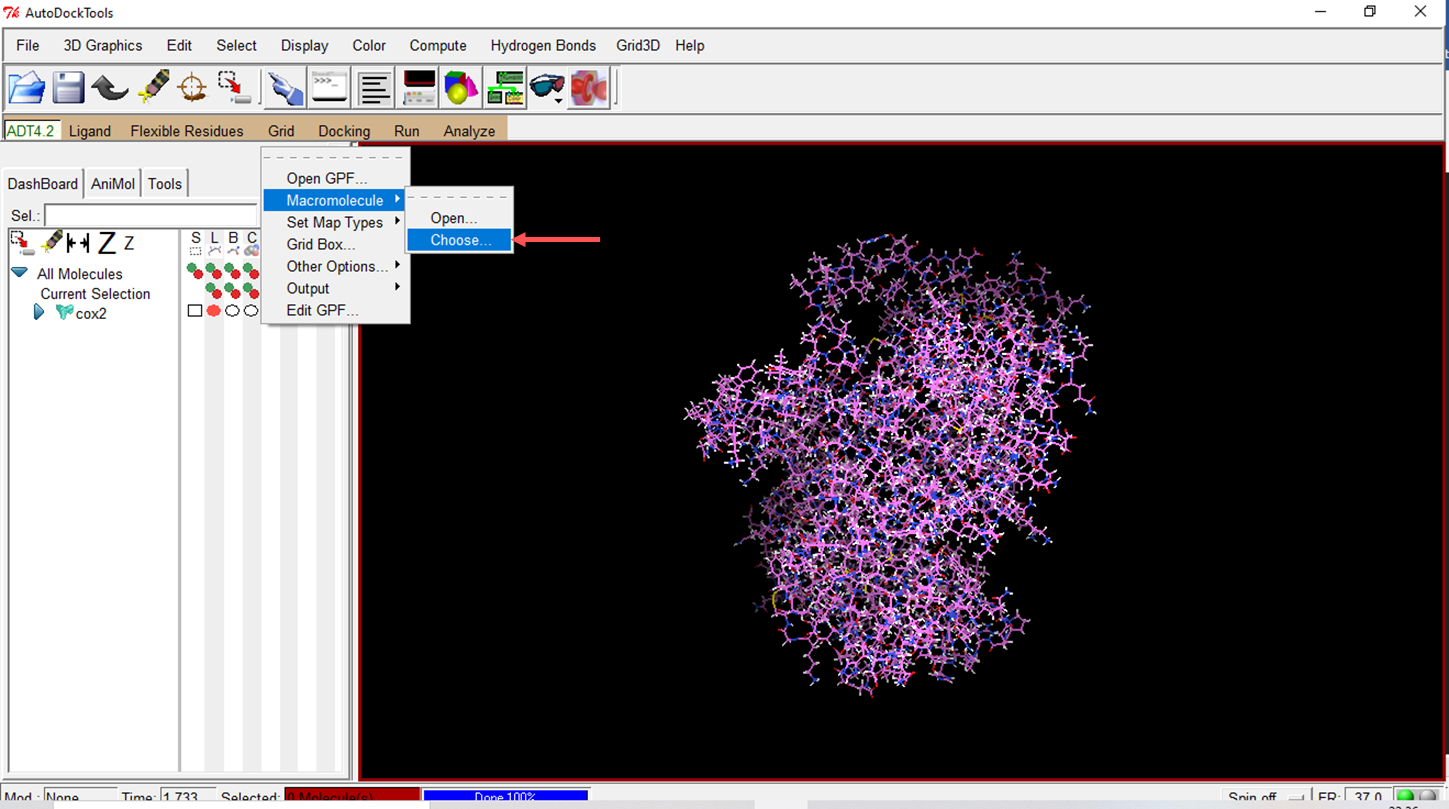
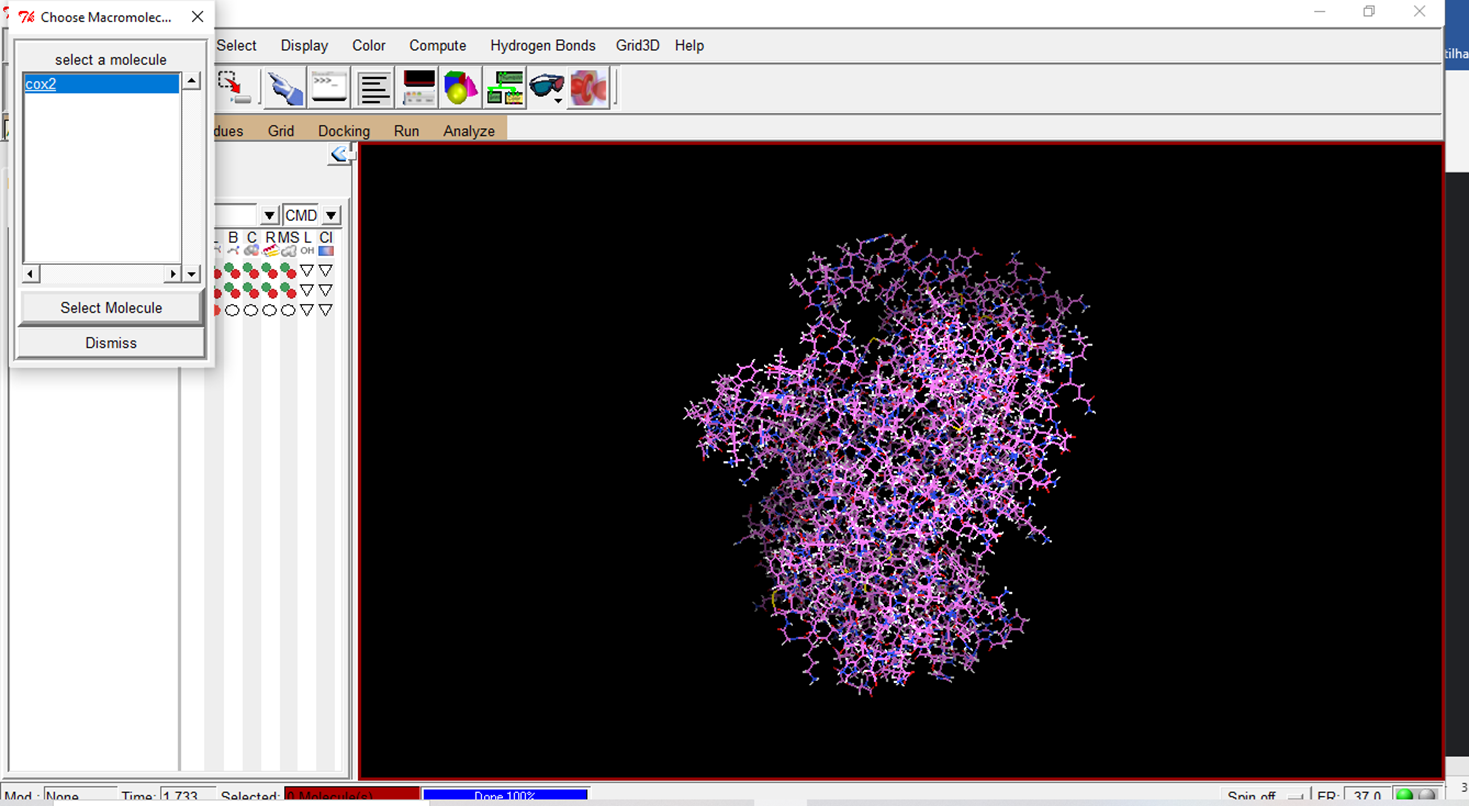
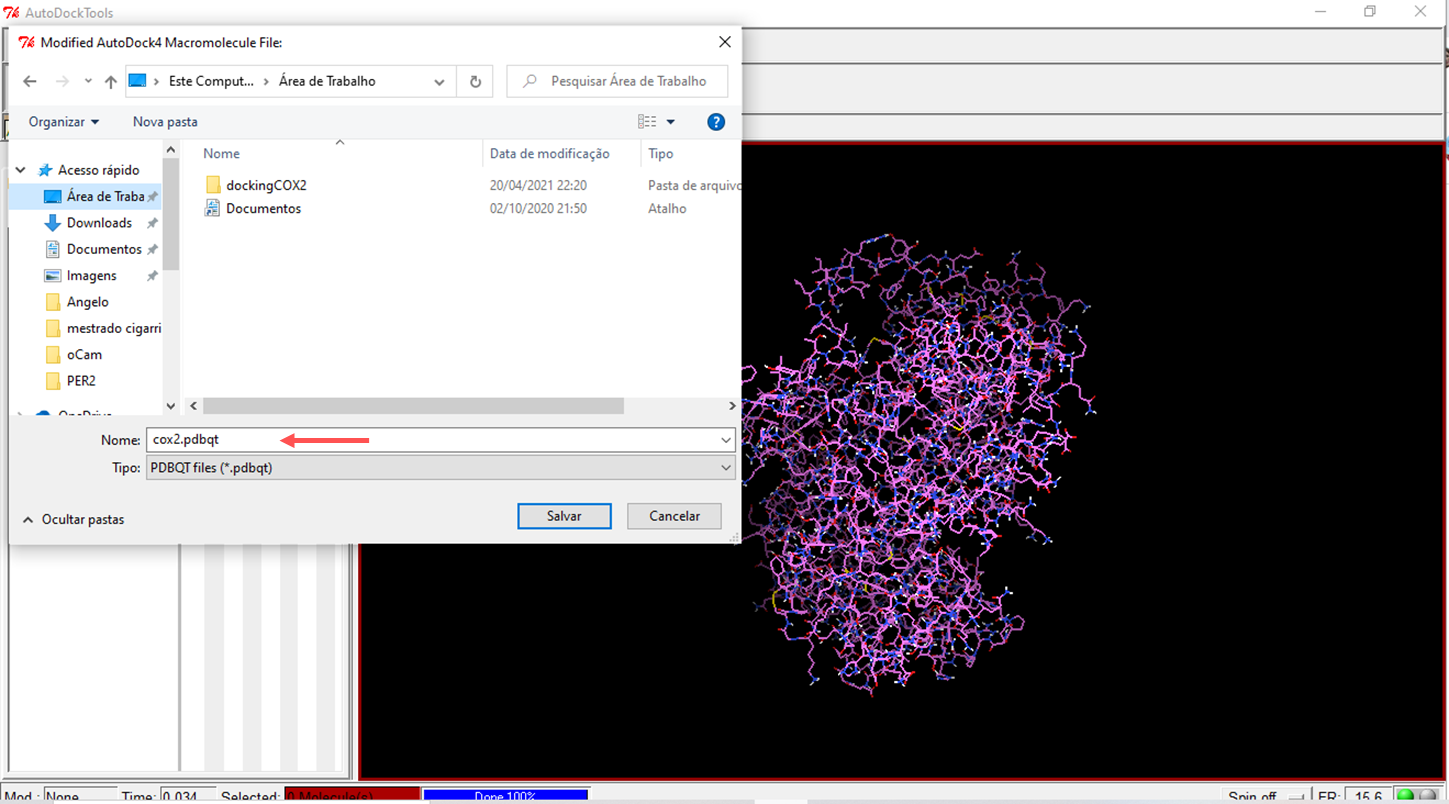
- Choose the prepared macromolecule to save
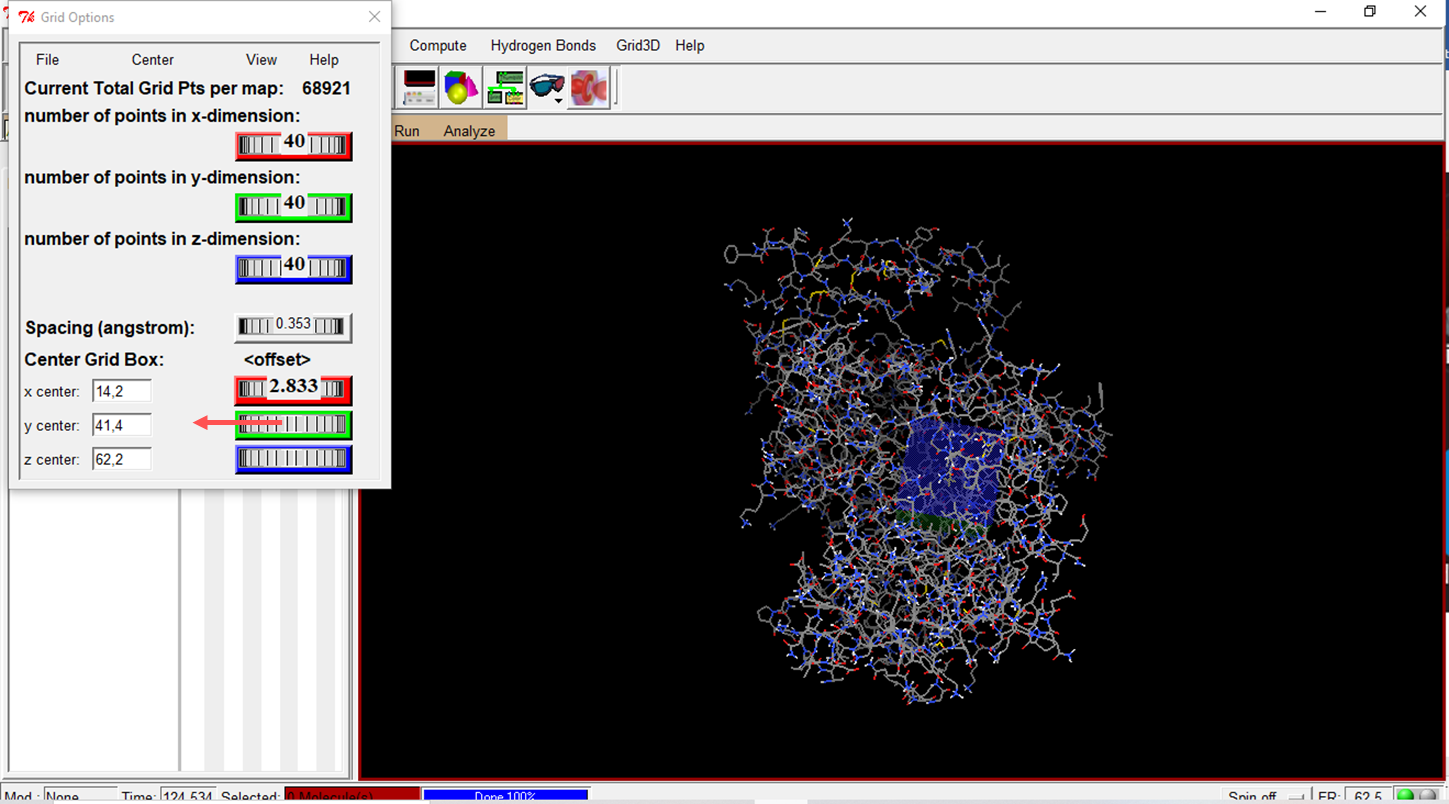
- Configure the Grid Box with the parameters provided by metaPocket 2.0
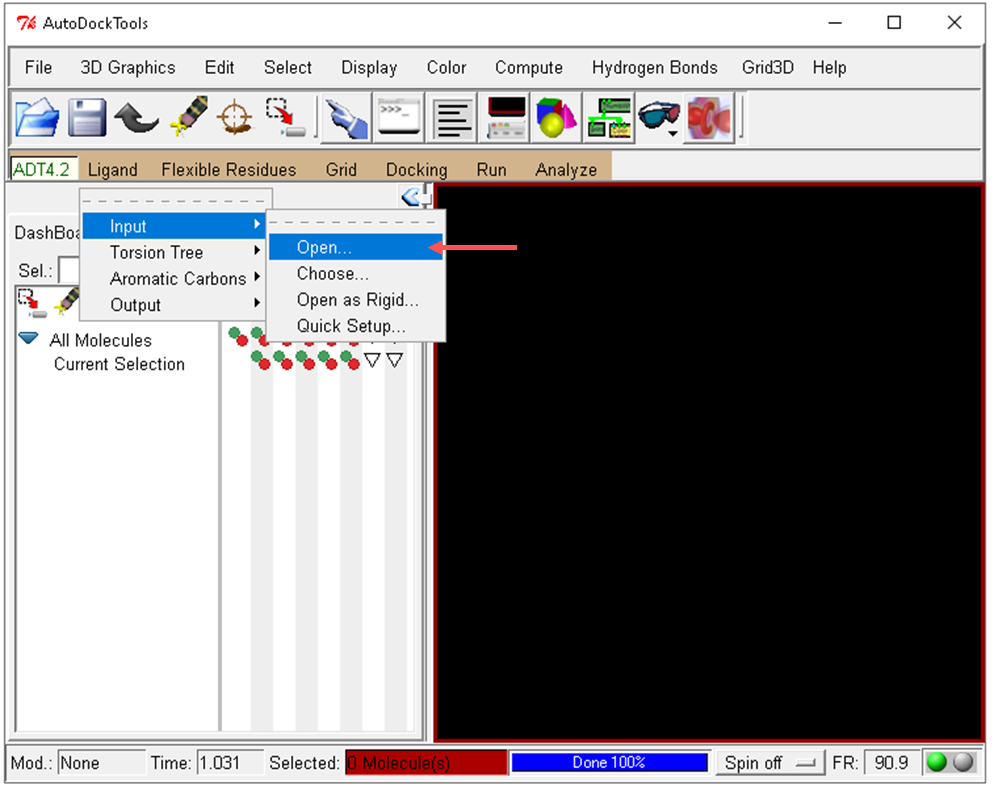
- Select the ligand
- Save the celecoxib molecule in PDBQT format
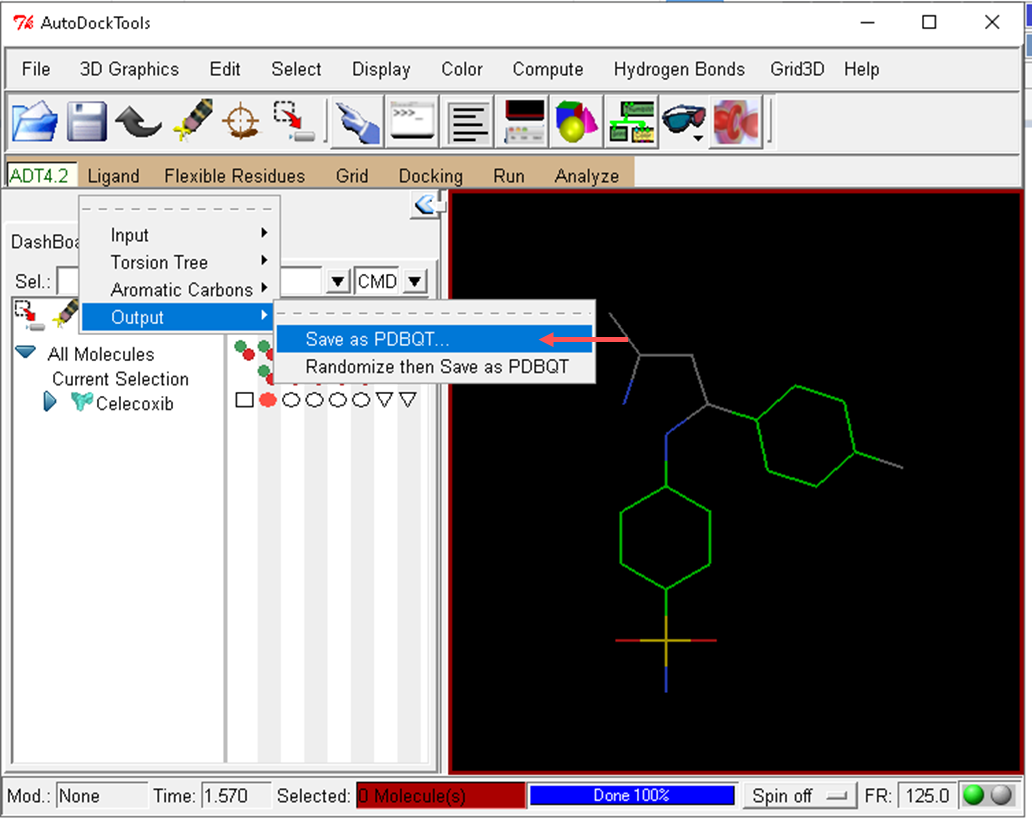
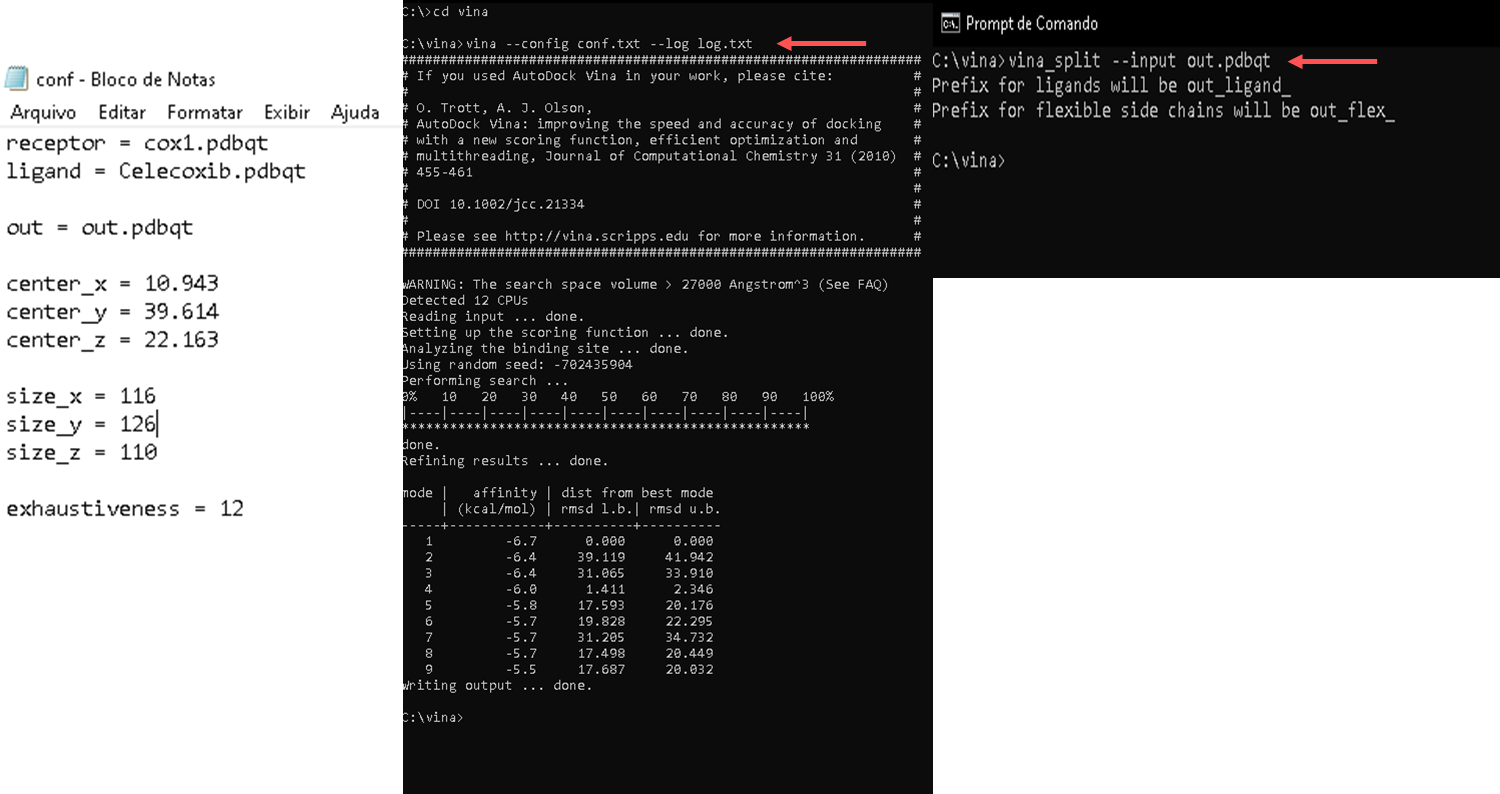
- Autodock Vina is molecular docking software and is generally used via the command line. The basic command to run Autodock Vina follows the following format:
vina --receptor <arquivo receptor.pdbqt> --ligand <arquivo ligante.pdbqt> --out <arquivo_saida.pdbqt>
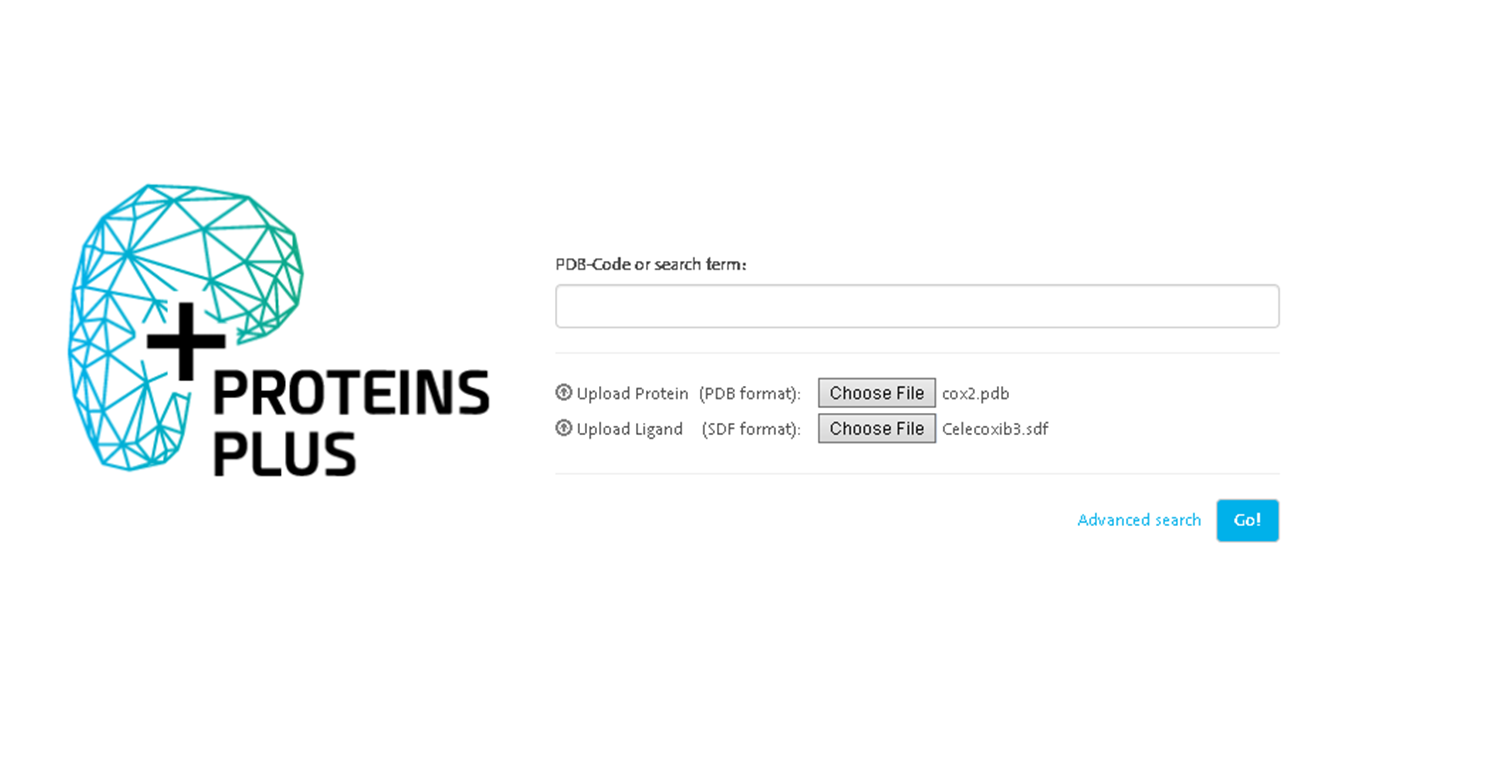
- Protein Plus The platform offers a series of integrated tools for protein and ligand analysis and visualization https://proteins.plus/
Choose the prepared files and click go.
- After loading the file and configuring the parameters, start the analysis by clicking the appropriate button (for example, "Submit" or "Run PoseView").
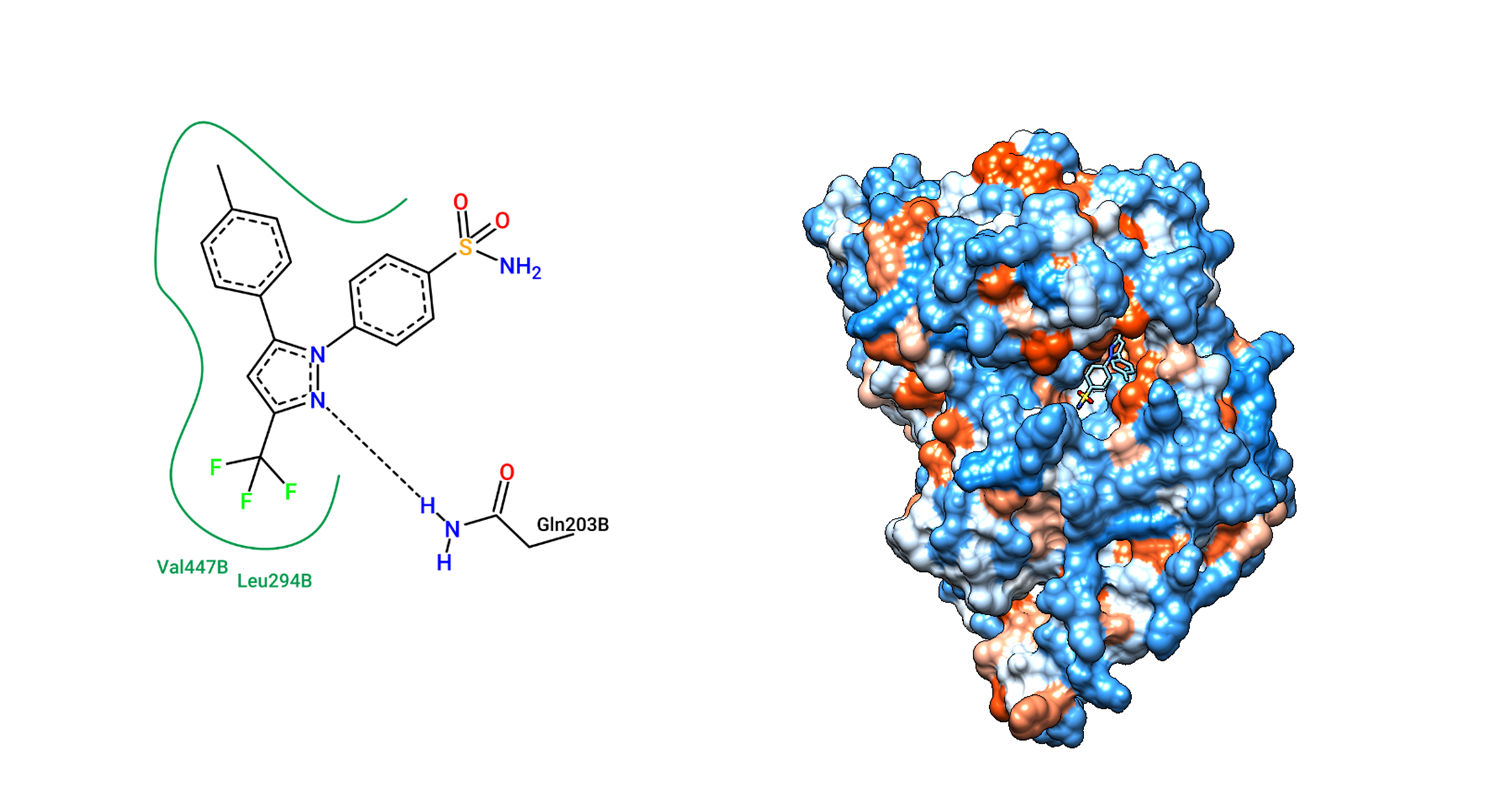
The tool will process the protein-ligand complex and generate a diagram of the interactions.
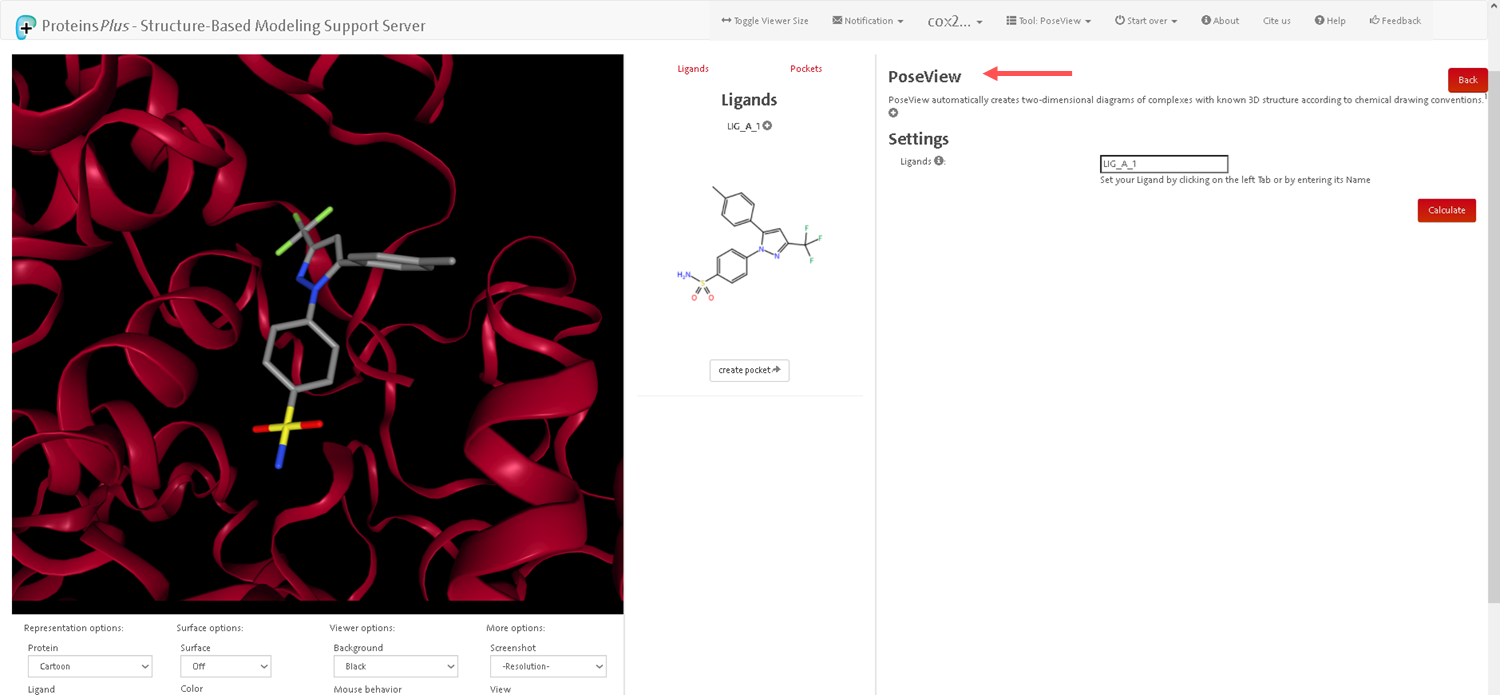
- After the analysis is completed, the tool will generate an interaction diagram. This diagram illustrates hydrogen bonds, hydrophobic interactions, and other important interactions between the protein and ligand.