Immune Saturation Genome Editing (Immune SGE): Variant Analysis in T cells
Nathan Camp, Sam Dawson
Saturation Genome Editing
Immune Cells
T cells
Primary cells
T cell receptor
CD3
CD28
CARD11
BCL10
IRF4
ssODN
CFSE
Abstract
This protocol details immune saturation genome editing (Immune SGE) and variant analysis in T cells. In brief, primary T cells are isolated from donor PBMCs, edited with a variant library centered around a guide cut site, and variants are scored based on how they affect T cell proliferation in response to T cell receptor (TCR) stimulation. This method works well for genes that regulate the response to TCR activation.
Steps
OVERVIEW: ssODN repair template and oPool design notes
Determine your editing region of interest, trying to stay within 20bp on either side of the cut site.
Add respective homology arms to both the 5’ and 3’ ends of the editing region such that ssODN is at least 140 bp long.
Introduce 3 synonymous mutations in the editing region within the guide homology region and the PAM. Ideally you have one mutation in the PAM site.
Use custom scripts to create your list of variants within the editing region.
Determine number of stop codons in library. Add up to 10 stop codons total if there aren’t enough.
Take all variants and add the homology arms (we do this in excel).
Add asterisks after the first 2 bases and before the last 2 bases if incorporating phosphorothioate bonds (this greatly improves stability as most cells contain nucleases that will degrade nonmodified ssDNA).
Get quote from IDT. Phosphorothiate and 3bp codes are not currently supported on IDT’s website when ordering oPools.
OVERVIEW: T Cell Culture method
Step 1 - Isolate CD4+ T cells from PBMCs
Step 2 - Activate CD4+ T cells with CD3/28 beads for 72 hours in Complete media + IL-2
Step 3 - Remove CD3/28 beads and allow cells to proliferate o/n
Step 4 - Transfect cells with Maxcyte
Step 5 - Collect day 2/3 samples for gDNA and RNA
Step 6 - Replace media with IL-2 every 2-3 days (M/W/F)
Step 7 - Take off IL-2 on day 9
Step 8 - Take 2-4e6 cells for gDNA and RNA
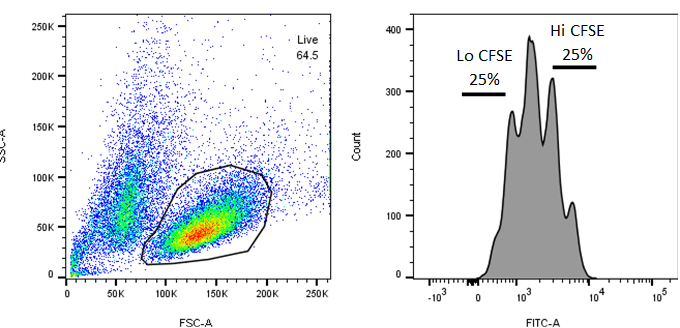
Step 9 - Stain cells with CFSE and stim with CD2/3/28 (no IL-2) for 72 hours
Step 10 - Sort cells on flow cytometer
Step 1: Isolate CD4+ T cells from PBMCs (starting from ~3.5 mL)
Pipette5mL of filtered FBS into a 50mL conical tube.
Thaw a vial of frozen cells in a 37°C water bath until cells are completely thawed.
Use a 5ml serological pipette to take cells from vial and slowly add the thawed cells dropwise into the FBS in the 50mL conical while swirling.
Add PBS 2% FBS (or EasySep buffer) dropwise to the 50 mL conical until the volume reaches 20mL.
Spin down cells and aspirate sup.
Optional step: If cell pellets appear red, add in 5mL of 1x ACK buffer. Let sit for 0h 2m 0s. Dilute out ACK with 20mL PBS before centrifuge.
Resuspend cells in 20mL CD4+ T cell isolation buffer.
At this point you can take 10µL of the cells for counting. While counting, spin down cells.
If you use hemocytometer to count total PBMC, make 1:10 dilution with Trypan blue before counting: If you use cell counter, follow manufacturer’s protocol. Record cell concentration (cells/ml) and calculate total PBMC counts.
Aspirate the supernatant and resuspend the cells to 5.0E+07cells/mL in EasySep buffer (PBS containing 2% FBS and 1millimolar (mM) EDTA).
Transfer cells to a 14 mL Falcon round bottom polystyrene tube.
Add the EasySep Human CD4+ T Cell enrichment cocktail at 50 uL/mL cells (e.g. add 100µLof cocktail to 2mL of cells). Mix well and incubate at Room temperature (15°C - 25°C) for 0h 10m 0s.
Vortex the EasySep™ D Magnetic Particles for 0h 0m 30s. Ensure that the particles are in a uniform suspension with no visible aggregates. (Solution should be uniformly brown).
Add the EasySep™ D Magnetic Particles at 100 µL/mL cells (e.g. for 2mL of cells, add 200µL of magnetic particles). Mix well and incubate at Room temperature(15°C- 25°C) for 0h 5m 0s.
Bring the cell suspension up to a total volume of 2.5mL (for 5ml tube), 5mL (for samples 2mL), 10mL (for samples 2mL) by adding EasySep buffer. Mix the cells in the tube by gently pipetting up and down 2 - 3 times. Place the tube (without cap) into the magnet. Set aside for 0h 5m 0s.
Pick up the EasySep™ Magnet, and in one continuous motion invert the magnet and tube, pouring off the desired fraction into a new 14 mL polystyrene tube.
- The magnetically labeled unwanted cells will remain bound inside the original tube, held by the magnetic field of the EasySep™ Magnet.
- Leave the magnet and tube in inverted position for
0h 0m 2s-0h 0m 3s, and then return to an upright position.
Take an aliquot for counting, and spin down.
Count and resuspend in cell culture media + IL-2 at 0.5e6 cells/mL.
Record the PBMC count and T cells count in the PBMC tracking file at the proper place.
Step 2: Activate CD4+ T cells with CD3/28 beads
Calculate volume of beads needed. We use 30µL beads/1e6 cells.
Vortex CD3/CD28 beads for 0h 0m 10s-0h 0m 15s to fully resuspend beads. Add desired volume of beads to 1.5 mL Eppendorf.
Add 500µL media into each Eppendorf tube and place in DynaMag™-2 magnet for 0h 0m 30s.
Remove media avoiding beads (should be visible held to the side of the tube).
Resuspend beads with media (same volume of beads suspension) and add to each well containing T cells.
Gently mix T-cells and CD3/CD28 beads with a P1000 pipette or 5ml serological pipette. Place cells in 37°C incubator for 72h 0m 0s.
Step 3: Remove CD3/28 beads
Observe cells under microscope before removing beads. Cells should be clumping in the well and most of cells should be visibly larger prior to removing beads.
Use a P1000 pipette or 5ml serological pipettes to gently pipet up and down 10 times to separate cells from beads.
Collect cells in 15 mL tube, at no more than 10 mL per tube (multiple tubes if culture volume is greater than 10 mL).
Place on magnet for at least 0h 1m 0s.
Carefully transfer cells in culture to a new tube without touching the beads.
Wash culture wells with 0.5mL-1mLfresh media and add to tube containing only the beads. Take the tube off the magnet to re-suspend the beads well.
Place on magnet for at least 0h 1m 0s to separate the beads and media, and then collect media to the tube containing cells.
Discard tube containing only beads.
Repeat steps 36 – 37 at least once more to ensure all beads are removed.
Count cells and record cell concentration as well as total cell numbers. Resuspend cells at a concentration to 0.5x106 cells/mL in fresh media + IL-2.
Culture at 37°C for ∼16 hours (16h 0m 0s) before transfection.
Step 4: Transfect cells with Maxcyte
If using crRNA, complex with trRNA. crRNA and trRNA are resuspended at 100micromolar (µM) and mixed 1:1. Add 15µL of crRNA + 15µL trRNA in sterile PCR tube, mix well. Heat to 95°C and allow to cool slowly to Room temperature,0h 30m 0s-1h 0m 0s.
Add 7.5µLof recombinant Cas9 (Cas9 stock is 40micromolar (µM)). You now have a guide:Cas9 ratio of 4:1. Allow mix to rest at Room temperaturefor 0h 15m 0s-0h 20m 0s.
While Cas9 guide complex is forming, spin down cells, resuspend in PBS and count. Spin down enough cells for transfection, then wash 1x with Maxcyte buffer.
Resuspend 5e6 cells in 82.5µL of Maxcyte buffer.
Add 5µL of oPool HDR library (oPool library is at 40 pmol/uL) to the Cas9 guide complex, then add cells.
Add cells to Maxcyte assembly and transfect with protocol “Expanded T Cell-1”.
Transfer cells to 4mL-5mL media + IL-2.
Step 5: Collect day 2/3 samples for gDNA and RNA
If there are enough cells (>10e6), collect 2 samples, each containing 2-3e6 cells. Spin down and aspirate supernatant.
For sample 1, isolate gDNA with favorite kit. We use the Quick-DNA miniprep plus kit from Zymo, cat. No. D4069.
For sample 2, resuspend in 600µL of Trizol reagent and store at -20°C or -80°C until processing.
Step 6: Replace media with IL-2 every 2-3 days (M/W/F)
Replace media with IL-2 every 2-3 days. If cell counts are <40e6, we plate cells at 1e6/mL. If cell counts are >40e6/mL (typically after 7 days post transfection) we plate at 2e6/mL.
Step 7: Take off IL-2 on day 9
1 day prior to CFSE labeling and restimulation with CD3/28, spin down cells and resuspend at 1-2e6/mL of media without IL-2.
Step 8: Collect day 10 samples for gDNA and RNA
Wash day 10 cells 1x with PBS. Resuspend in 10mL PBS and count cells.
Like section "Collect day 2/3 samples for gDNA and RNA" above, collect 2 samples, each containing 2-3e6 cells. Spin down and aspirate supernatant.
For sample 1, isolate gDNA with favorite kit.
For sample 2, resuspend in 600µL of Trizol reagent and store at -20°Cor -80°Cuntil processing.
Stain 80e6 cells with CFSE and stim with CD2/3/28 (below).
Step 9: Stain cells with CFSE and stimulate with CD2/3/28 (no IL-2) for 72 hours
Resuspend each vial of CFSE (Thermo, cat. No. C34570) in 18µL DMSO.
Resuspend 80e6 cells in 40mL PBS. Add 20µL of CFSE, mix well and incubate at Room temperature for 0h 5m 0s.
Add 5mL of FBS, mix well and incubate at Room temperature for 0h 5m 0s.
Spin down cells, wash 1x in10mL media without IL-2.
Resuspend cells in 40mL media without IL-2 and add 125µL of ImmunoCult Human CD2/3/28 (Stemcell, cat. No. 10970). Place in incubator and wait 72h 0m 0s-96h 0m 0s before sorting.
Step 10: Sort cells on flow cytometer
72-96h 0m 0s post stimulation, spin down cells and resuspend in PBS at 5-10e6 cells/mL.
Isolate gDNA from both CFSE Lo and CFSE Hi fractions.