Human Liver Core-Needle Biopsy Processing Protocol
Catia Perciani, Ian McGilvray, Sonya Macparland, Diana Nakib
Abstract
Dissociating and isolating cells from a liver biopsy for single cell RNA-sequencing and storage for additional multiomic applications. We have developed a high-thoughput method of dissociating liver tissue for transcriptomic analysis, as well as preparing sections for histological, immunohistochemical and functional assessment.
Steps
Buffers and Solutions
Biopsy collection tube:
Add 10 mL HBSS 1X to a 50 mL tube and keep it on ice or in the fridge for the liver biopsy collection (label the tube with the sample ID and date). All catalogue numbers in materials.
PBS-0.04% BSA:
Add 100 µL of 2% BSA (same as 20 mg/mL) to 4.9 mL of PBS.
PBS-5% FBS:
Add 2.5 mL of FBS to 47.5 mL of PBS.
Enzyme Master Mix:
Add 50 µL collagenase + 50 µL protease to 0.4 mL of HBSS 1X.
Tissue Assessment and Allocation
Transfer the biopsy to a petri dish and examine its size and whether it is possible to split it into 2 sections: one half for tissue dissociation and another half to then be split into two or three and be stored as 1) snap-frozen, 2) in OCT and/or 3) in formalin.
Tip: Tip: Place the petri dish on a flat ice pack to section the biopsy while on ice
Using the disposable scalpel and forceps, measure and the biopsy for dissociation and storage according to the below measurements:

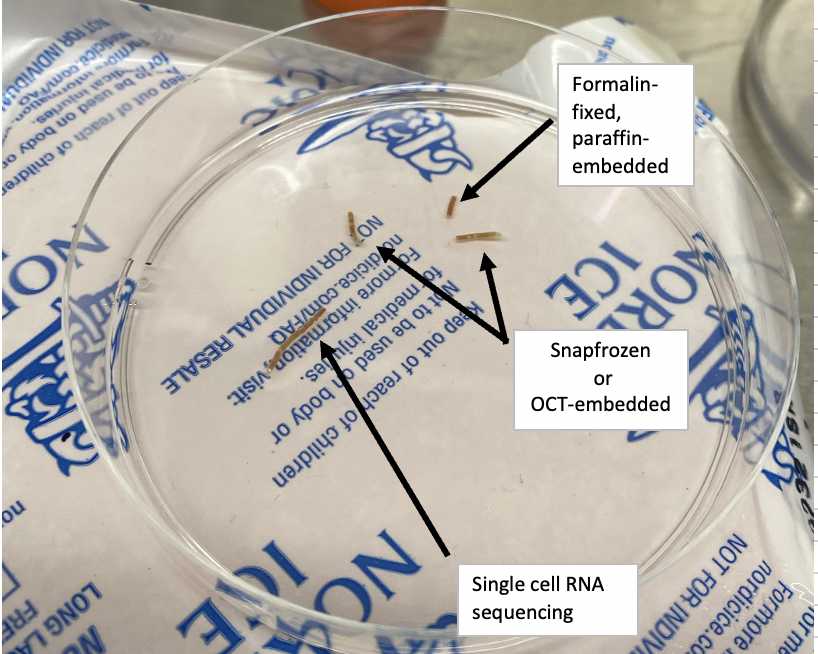
For tissue dissociation/scRNA-seq:
Minimum biopsy length of ~1.0 cm should be allocated.
For snap-freezing/OCT-embedding/formalin-fixing:
Minimum biopsy length of ~0.5 cm for each storage method.
Methods for snap-freezing, OCT-embedding and formalin-fixing are described in Human Liver Tissue Storage Methods For Multiomic Applications protocol.
Biopsy Dissociation
Transfer the biopsy section to be dissociated into a 1.5 mL Eppendorf tube containing 200 µL of enzyme mix kept at 37ºC.
Using a sterile spring scissor cut the biopsy into as many pieces as possible. 1 x sterile spring scissor (14 mm cutting edge, 0.275 mm tip diameter, 12 cm length, Fine Science Tools)
Secure the Eppendorf tube inside a petri dish and place it in an incubator (37ºC) on a shaker set at 500 rpm for 10 min.
500rpm
Examine for the presence of undigested fragments.
If there are undigested fragments, repeat Steps 8 and 9 to improve biopsy dissociation and digestion.
Filtering and Washing of Cells
Filter the cell homogenate using a 70 µm cell strainer on a 50 mL conical tube or through the strainer of a 5 mL round-bottom FACS tube.
Tip: Use a 20-200 µL pipette to transfer the cell suspension to central point of the cell strainer. Tip: Use a 20-200 µL pipette to transfer the cell suspension to central point of the cell strainer.
Wash the cell strainer with 3ml of PBS-5% FBS.
Spin the cell suspension at 400g for 5min at 4ºC.
400x g,4°C
Remove the supernatant.
Red Blood Cell Lysis
If the pellet is Pink/Red in color:
Treat the cells with 1 mL of ACK lysing buffer for 5 minutes at room temperature.
0h 5m 0s
Room temperature
Add 3ml of PBS-5% FBS.
Spin cell suspension at 400g for 5min 4ºC.
400x g,4°C
Remove supernatant.
Submitting Sample for Single-cell RNA Sequencing
Resuspend the pellet in 100ul of PBS-0.04% BSA.
Transfer the cell suspension to a 1.5ml Eppendorf tube and keep the cells on ice for processing for 10x Genomics scRNA-seq.
On ice
Left over cells can be counted, spun down with the below specifications, and resuspended in 1ml of Freezing Media (10% DMSO in FBS) and placed in a cryovial for storage.
400x g,4°C
Wrapping up
After completing the protocol, make sure to clean the scissors and send it for autoclaving (cover the sharp part with a 1.5 mL Eppendorf tube and secure it with an autoclave tape).

