Highly Parallel Droplet Sample Preparation for Single Cell Proteomics
Andrew Leduc, Richard Huffman, Joshua Cantlon, Saad Khan, Nikolai Slavov
single-cell proteomics
mass-spec sample preparation
sample preparation
high-throughput sample preparation
nPOP
nano-ProteOmic sample Preparation
droplet sample preparation
Abstract
Protocol for preparing single cells for mass-spec analysis by nPOP as described by Leduc et al., 2021, 2022 DOI: 10.1101/2021.04.24.441211. nPOP uses piezo acoustic dispensing to isolate individual cells in 300 picoliter volumes and performs all subsequent preparation steps in small droplets on a fluorocarbon-coated slide. This design enables simultaneous sample preparation of thousands of single cells in a single batch. This includes lysing, digesting, and labeling individual cells in volumes below 20 nl. nPOP supports different experimental designs including label free analysis, TMT 18plex with carrier, mTRAQ, and TMT-10plex for TOF instruments (see steps).
Data, resources and additional information: https://scp.slavovlab.net/Leduc_et_al_2022 and https://scp.slavovlab.net/nPOP
Andrew Leduc presented a detailed workflow of nPOP at the 4th Single Cell Proteomics Conference available on YouTube: https://youtu.be/DJ1U_KpMNcY
The protocol uses the CellenONE system for liquid handling and cell sorting and assumes basic level of familiarity and training on the CellenONE. If there are any questions, please email leduc.an@northeastern.edu
Steps
Setting up system
This protocol enables highly paralleled single cell sample prep for both label free analysis and multiplexed analysis.
We provide options for label free, TMT 18 plex with carrier, mTRAQ labels for multiplexed DIA analysis, and TMT 10 plex labels for isobaric multiplexing on TOF instruments.
Which option you choose depends only on the folder of droplet layout files you use. As well as skipping all sets involving labeling and quenching for LF analysis.
To get started with nPOP, you will need to move the the files from this repository into your CellenONE. There are three important classes of files contained:
- Tasks (discrete actions)
- Runs (made up of tasks)
- Designs (droplet layouts)
To install these files on your instrument, navigate to the following base directory (this is what it is on my instrument, at least):
C: > Users >Scienion AG > CellenONE
- Droplet layouts go in the "Pattern" subdirectory
- Run-associated files go in the "Settings/Run" subdirectory
- Task files go in the "Settings/Task" subdirectory
Close the CellenONE software and reopen to verify that the files have appeared in the corresponding locations (run-associated files in the "Run" panel in the Main tab, tasks in the "Do task" sub tab of the nozzle setup tab, and droplet layouts can be loaded in as field files).
For further clarity, link for label free. Link for 10 plex TMT on TOF. Link for TMT 18 plex with carrier (14 cells per set).
Link for plexDIA.
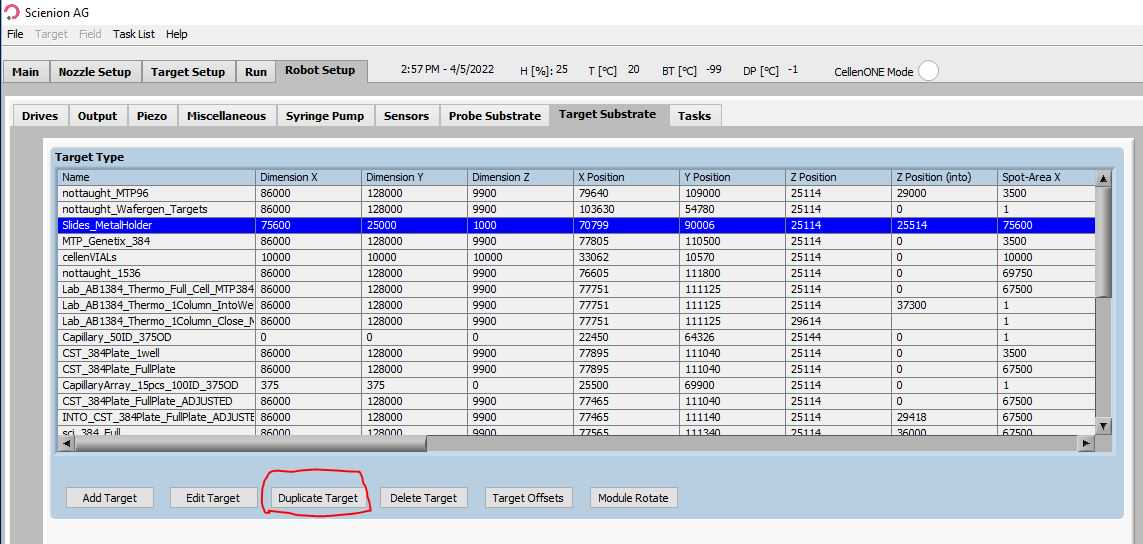
Next, we need to make one more target for aspirating samples off of the surface of the glass slide.
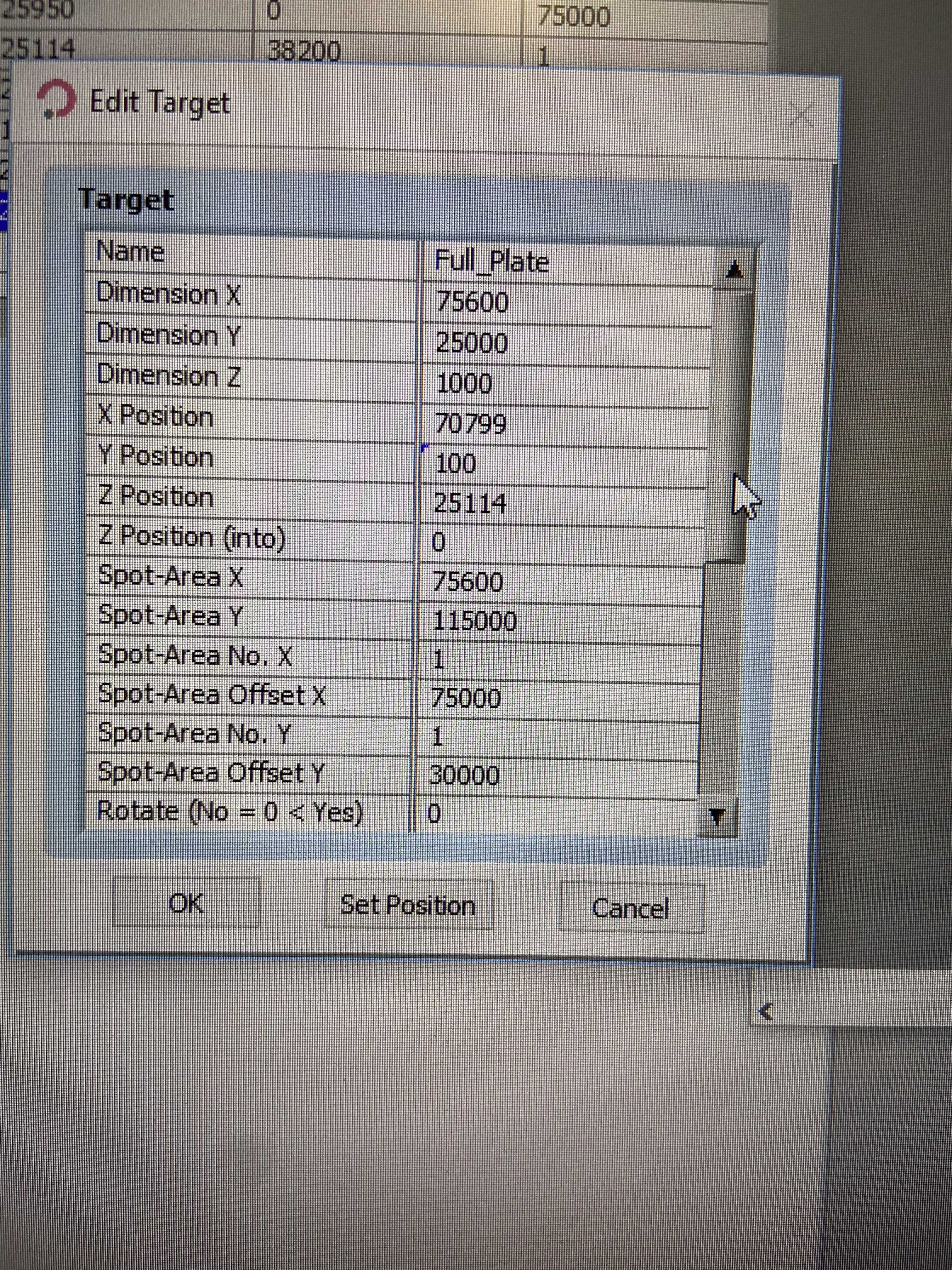
This should be the exact same as the last one you just made (so make a new duplicate of "Full_Plate" and call it "Full_Plate_SlidePickup") but the Z Position should be ~29000. This is different on each instrument, so what you can do is go to the Drives tab, move the nozzle to the "Slides_MetalHolder" Target area, and inch down the z height so that it is just slightly off the surface of the slide. Once satisfied, set the new value for Z position as you did in step 3.
Time to test our new setup! In the main tab, select your Target area to be "Full_Plate" and the run to "SpotRun". Then load the following Field file: System_setup > Setup_slide_all.fld. This method should place a small droplet in all 4 corners of each slide. If you're not happy with droplet positions, you can adjust the spacing in between slides with the parameter shown below. After changing it, be sure to hit ALL to apply the spacing to for the distance between each slide. Once this is set, you will need to apply these settings to all the field files that you use (load everything and save them with the changes you made to these parameters).
What you need to get started
To get started, you will need a few things. These consumables can all be purchased from Scienion
Personally, I have had the most success operating with two PDC nozzles from Scienion:
- A regular glass PDC, size medium, for handling cells
- A PDC 70 with coating type 2. This is for handling organic solvents such as DMSO and protein mixtures.
For slides, we use the H1 slides which are regular glass slides that have a flourocarbon coating.
In terms of the sample prep itself, you will need your cells (obviously), DMSO, Trypsin Gold, Benzonase endonuclease, HEPES pH 8.5, any labeling reagents (TMT, mTRAQ, etc.), and triethylammonium bicarbonate (TEAB) if using mTRAQ.
Once you have everything you need to get started, it's worth investing some time to get your workflow ironed out prior to attempting a full prep. In the following two sub-points you will find some preparatory steps that I take alongside some troubleshooting suggestions.
Getting the nozzles in the exact same condition each time you start a prep.
My routine is to use the syringe attachment to clean and flush the nozzle and lines within the PDC. I place the tip of the PDC in water that is sonicating and pull water in and out of the PDC for ~5 minutes, followed by a minute of 100% ethanol and then another minute of water. Be careful not to get the black ring of the PDC wet, as this can cause electronics to short if you use directly after.

Practice with the reagents you will be using before starting a prep.
Several factors such as humidity, room temperature and the specific nozzle can have large effects on dispensing accuracy.
As an example, for dispensing mTRAQ labels, I use DMSO as I found it easiest to work with, however, we have also used Ethanol and IPA for dispensing mTRAQ in the past. This can work well, but, requires you to aspirate air and then ~20 uL of organic solvent prior to aspirating label and dispensing.
Some tips that have worked for me:
-
the trypsin is highly viscous, so be sure to run "dip wash" after aspirating, and reduce frequency, voltage, and pulse considerably
-
if a droplet seems to be shooting off to the side, dabbing the bottom of the tip twice with a kimwipe (damp with ethanol) can really help. If the issue persists, move nozzle to home position, turn on continuous dispensing, and try cleaning tip with an ethanol-dampened kimwipe for ~60 seconds.
-
Generally higher voltage has helped with DMSO dispensing. Also make sure humidity is not too high when dispensing DMSO, this can cause droplets to stick to the end of nozzle.
Carrier and Reference Preparation (***Only if Carrier is used***)
Prepare cell pellets of at least 500,000 cells for all relevant cell types . Add 100% DMSO to cells to a cellular concentration of 6000 cells/ul. Incubate cells in DMSO for 20 minutes to lyse cells. Add mass spectrometry grade water to bring solution to 2000 cells/ul. More information on carrier and reference preparation is available at Petelski, A.A., Emmott, E., Leduc, A. et al. Multiplexed single-cell proteomics using SCoPE2. Nat Protoc 16, 5398–5425 (2021).https://doi.org/10.1038/s41596-021-00616-z
Add 1x benzonase, 100mM TEAB and 20 ng/uL of trypsin. Digest sample at 37 degrees C overnight.
Label carrier and reference samples with desired labels. Dilute samples before combining such that the composite sample is at 200 cells/uL of carrier and 5 cells/uL of reference.
Preparing CellenONE
In the target spotting location, place the glass slide holder (make sure it fits well) and then place your fluorocarbon-coated glass slides (CellenONE H1 Slides) in their designated slots (make sure they are all level).
In the probe location, place any desired 384 well plate for storing and pickup of cell suspensions and reagents used in sample preparation. On the CellenONE "Main" tab, set the probe location to the relevant 384 well plate and set the target location to our new "Full_plate".

Preparing cell suspension
Suspend cells in 1X PBS at a concentration of 200-300 cells/uL. If cells are prone to clumping, first filter with a 40 micron filter to ensure a fast pace of cell sorting.
DMSO for Cell Lysis
Turn the humidifier on and set it to maintain 60% relative humidity and set the plate temperature to maintain 19C.
Ensure that "Volume Entry is selected in the field settings and that the "DMSO_L" field file is selected.
On the main page, set the Run to "Sample_pickup_and_dispense".
Make sure the second nozzle (the type-2 PDC is selected).
Load 20 uL of 100% mass-spectrometry-grade DMSO into all relevant wells in the 384-well plate. The number of wells you need to use depends on the sample prep you are doing (wells used at each step can be found in the field file). For simplicity, you can also just load wells D1-D8 as this will work for any layout.
Make sure the plate is secure, and run the "Sample_pickup_and_dispense" method!
DMSO droplets for lysis should be automatically dispensed over the relevant slides. After run is over, check images to make sure droplets did not deviate (these can be found in the Run folder you created/selected when starting prep).
If deviations are noticed, wipe droplets on the relevant slide with a KimWipe, flip slide over, and re-dispense for required slide. When removing slides, be careful not to move or touch adjacent slides as this will cause misalignment of droplets for subsequent steps and likely failed cells/preps for those drops.
Cell Dispensing
Change the system to "Droplet" mode by unchecking "Volume Entry".
Load the "Cells" field file and ensure spots correspond to one droplet.
If you are dispensing multiple separate cell suspensions, remove a portion of spots, (remove some from each cluster if using labels) to ensure analysis of all relevant cell types.
We also suggest leaving some droplets without any cell to serve as negative controls.
On main tab, set run program to "CellenONE_Basic".
Aspirate 10-20 uL of your cell suspension from the 384-well plate using the leftmost PDC (the glass one).
Dispense first cell type.
Repeat steps 17 and 18 for additional cell types
Evaporation Control
After cell dispensing is finished, set humidity control to 75 % relative humidity and set cooling control to "dew point chase" with a deviation of -.75 degrees.
Change to "SpotRun" in the Run menu.
Load the perimeter field file.
Dispense a perimeter of system water to control local evaporation.
Start preparing digest.
Digestion
To prepare the digest you will need, Trypsin Gold, Benzonase, and HEPES pH 8.5.
First prepare 60 uL of a solution that contains Trypsin gold at 120 ng/ul, 5mM HEPES, and .5 x Benzonase.
Place in a 1.5 mL tube.
This step is optional based off how easy dispensing trypsin solution is for user. In this step we degas the trypsin solution on ice. Find a bottle with the same opening as the cellenONE bottle where the system water is stored. Fill with ice to brim and place in a styrofoam container filled with ice.
Then place trypsin in ice (cap open), attach vaccum attachment used for degassing cellenONE water, turn on vacuum, and wait for 10 minutes.

How much master mix you aspirate depends on how many cells you are preparing. If preparing 2000 cells using the 14 cell per cluster method, we advise:
-
Fill 2 wells of the 384-well plate each with 30 uL of your master mix
-
Aspirate 20 uL of master mix with the PDC type-2 out of each, one after the other so the PDC contains 40uL of master mix in total.
-
Make sure to run dip wash immediately after aspirating master mix to ensure stable droplet.
Set Run file to "SpotRun" and load the "Digest" field file.
Dispense master mix.
Let single cell digestion proceed for 4 hours.
To control evaporation, you can reload the "Perimeter" field file and refresh perimeter field with system water 1-2 times throughout digestion depending on how much the perimeter seems to be evaporating.
Either PDC should work fine for this.
After digestion turn off humidity and cooling and wait ~15 minutes and let single-cell peptides dry out on the slide.
This is a pause point. Labeling can proceed the next day as peptides rest dried out on the slide.
If doing label free, proceed to the "Prepare 384 well plate for Storing Sample" section. The pickup of all 384 samples will take nearly a full day, so you can leave this until the following day if desired.
Labeling
The labeling routine depends a bit on the labels that you are using. We only have experience with TMT and mTRAQ.
For TMT we have found the labeling proceeds best in pure organic phase. For mTRAQ, sufficient and well buffered aqueous phase is needed.
If using mTRAQ, Load the field file for "ReHydrate" and dispense 200 mM TEAB to each spot. Also be sure to set cooling and humidity settings back to as previously set during digest.
If multiplexing reagents are stored in acetonitrile, evaporate off acetonitrile completely in speed vacuum on low heat or lyophilizer.
Resuspend TMT labels in 100% DMSO at a concentration of 1/3 maximum strength.
Critical: Pipette mix labels vigorously to prevent TMT crystals from forming as these can interfere with label dispensing.
Load all labels into wells G1 --> G# of labels you are using.
Load the field file for the first Label. Set the run method to "SamplePickup_And_Dispense" and run labeling.
I suggest leaving the light on and the camera so you can make sure there is no residual DMSO on the tip of nozzle.
If residual DMSO is present, you can pause the run with the "nozzle setup" button and dab tip with a Kimwipe to remove any hanging droplets.
Critical: Check droplet stream before and after dispense to ensure the label was accurately dispensed as there will be no imaging validation until end. Imaging each field after each label will significantly increase time for dispensing labels.
Quenching Labeling Reaction
Turn on humidity and cooling to previous settings used during digest.
Prepare a solution of 5% hydroxylamine (HA) diluted with mass spectrometry grade water.
Load the HA field file and set the run method to "Spot_Run".
Load 30 uL into two wells of the 384 well plate. Aspirate 40 uL of HA by taking 20 uL from each well.
Dispense HA and incubate for 20 minutes.
Repeat step 34 for two quenching steps.
Prepare 384 well-plate for Storing Sample
Pipette desired carrier amount diluted in 2 uL cells worth of combined carrier and reference into the wells of your 384-well plate. For labeled methods this will be wells J1-O24, again, this depends on method being used and so you can reference field file to determine which wells need to have carrier.
If not using carrier, fill every well with 2 ul of mass-spec-grade water.
Sample collection
Remove one of your nozzles from the CellenONE.
Change the target substrate in the main tab to "FullPlate_SamplePickup" that we set up in beginning. Change Run to "Pickup_off_Slide". (for LF or 3plex, use the "Pickup_off_Slide_smallVol" run)
Fill your wash tray with acetonitrile. Run the "Sample pickup" method. Watch to make sure the first few go well. You will likely need to refill the wash tray with ACN once or twice throughout the pickup, so check back in an ~1 hour.
Dry down each sample in a vacuum evaporator (we use a SpeedVac) for 30 minutes to remove any residual DMSO in the samples and store in -80 until samples are ready to be run on LC/MS.