High-throughput single-cell transcriptomics of bacteria using combinatorial barcoding
Karl D. Gaisser, Sophie N. Skloss, Leandra M. Brettner, Luana Paleologu, Charles M. Roco, Alexander B. Rosenberg, Matthew Hirano, R. William DePaolo, Georg Seelig, Anna Kuchina
single-cell transcriptomics
combinatorial barcoding
microbial SPLiT
high-throughput RNA sequencing
Gram-negative and Gram-positive bacteria



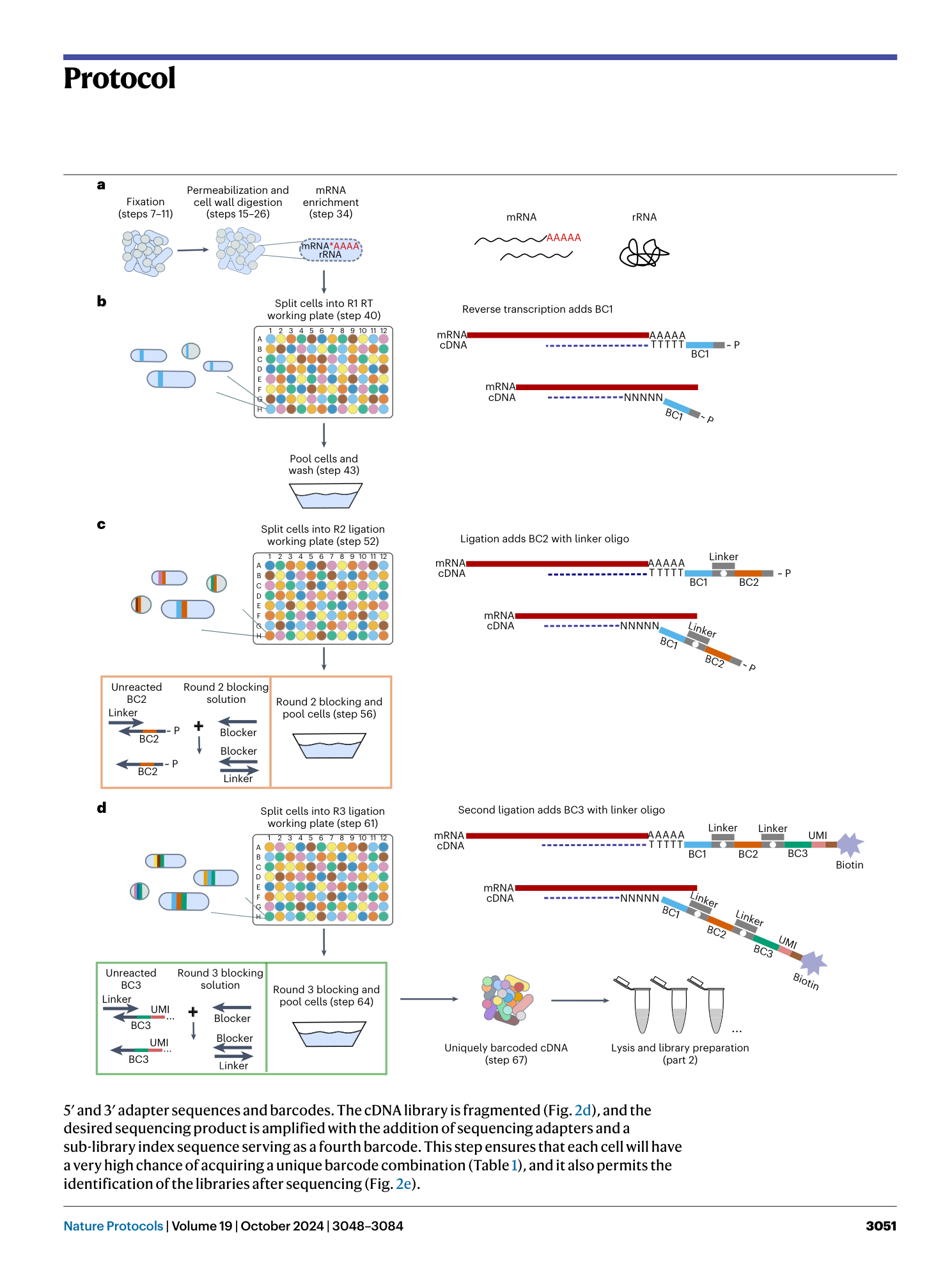
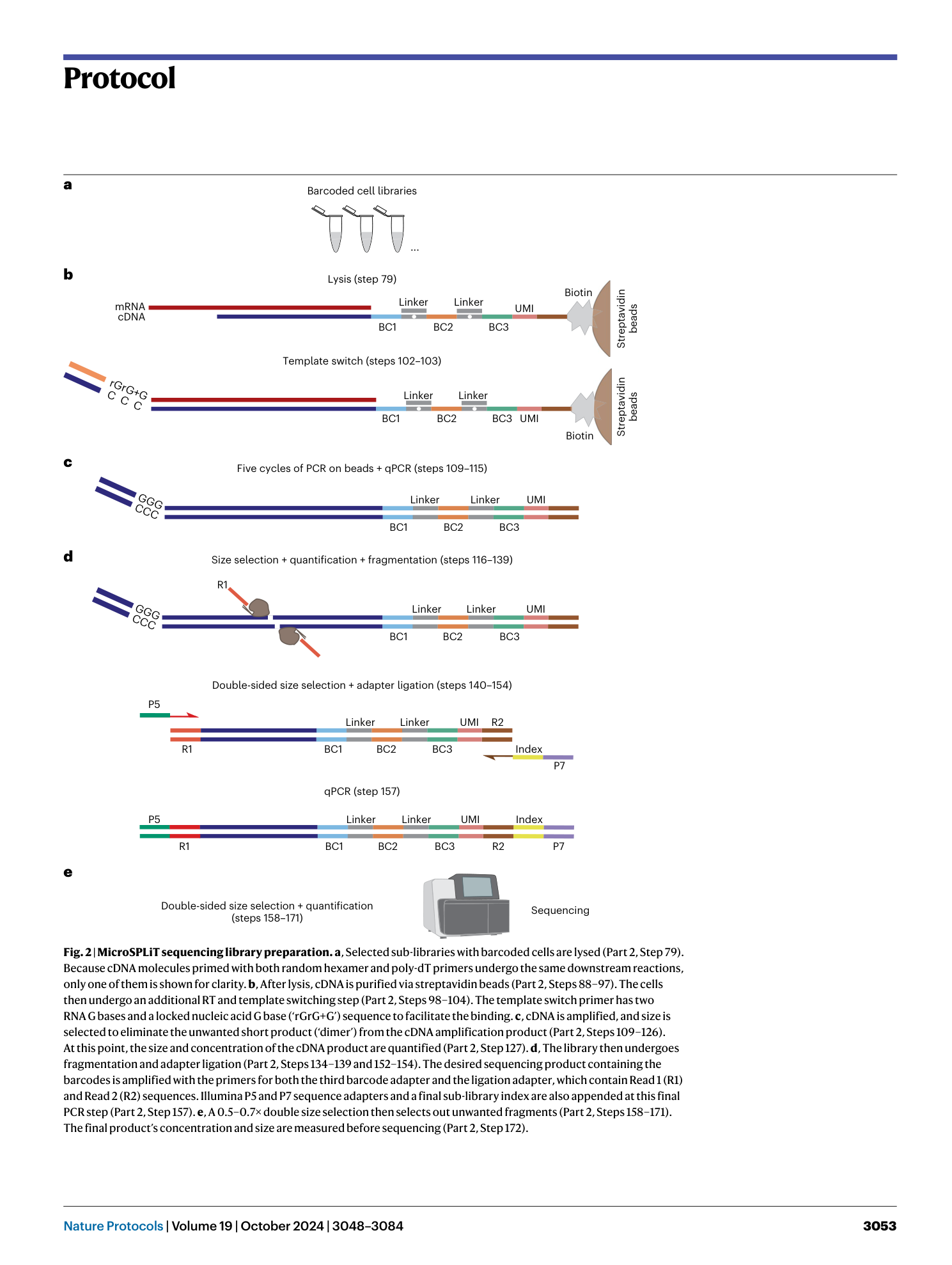


















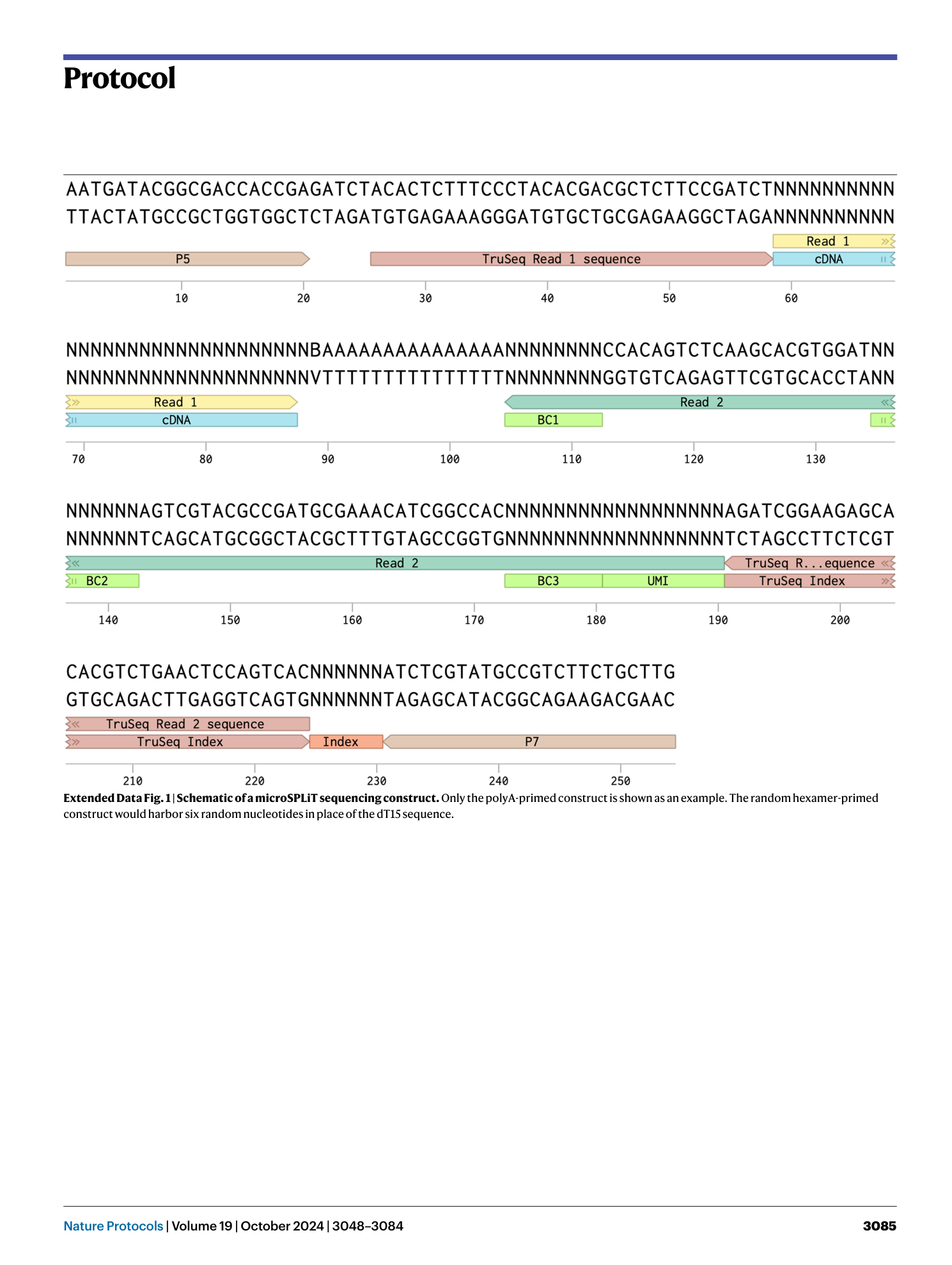
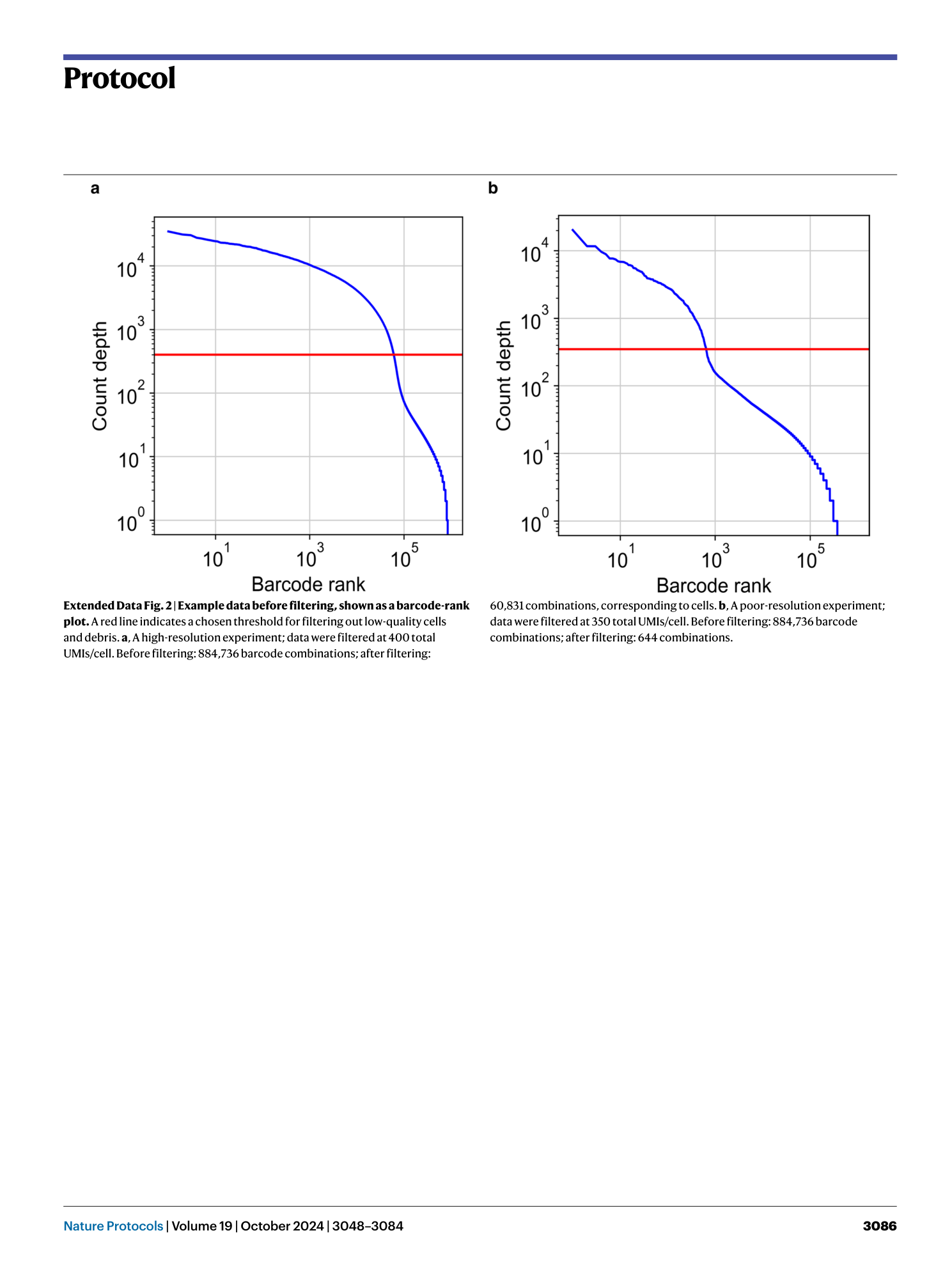



Extended
Extended Data Fig. 1 Schematic of a microSPLiT sequencing construct.
Only the polyA-primed construct is shown as an example. The random hexamer-primed construct would harbor six random nucleotides in place of the dT15 sequence.
Extended Data Fig. 2 Example data before filtering, shown as a barcode-rank plot.
A red line indicates a chosen threshold for filtering out low-quality cells and debris. a , A high-resolution experiment; data were filtered at 400 total UMIs/cell. Before filtering: 884,736 barcode combinations; after filtering: 60,831 combinations, corresponding to cells. b , A poor-resolution experiment; data were filtered at 350 total UMIs/cell. Before filtering: 884,736 barcode combinations; after filtering: 644 combinations.
Supplementary information
Reporting Summary
Supplementary Tables 1–4
Supplementary Table 1: cost estimate of microSPLiT. Supplementary Table 2: cost estimate and comparison of high-throughput bacterial scRNA-seq methods. Supplementary Table 3: sequences of oligonucleotides and source barcode plates used in microSPLiT. Supplementary Table 4: barcode sequence files for STARsolo analysis.

