Hi-C 3.0: Improved Protocol for Genome-Wide Chromosome Conformation Capture
Denis L. Lafontaine, Denis L. Lafontaine, Liyan Yang, Liyan Yang, Job Dekker, Job Dekker, Johan H. Gibcus, Johan H. Gibcus
Abstract
The intricate folding of chromatin enables living organisms to store genomic material in an extremely small volume while facilitating proper cell function. Hi-C is a chromosome conformation capture (3C)-based technology to detect pair-wise chromatin interactions genome-wide, and has become a benchmark tool to study genome organization. In Hi-C, chromatin conformation is first captured by chemical cross-linking of cells. Cells are then lysed and subjected to restriction enzyme digestion, before the ends of the resulting fragments are marked with biotin. Fragments within close 3D proximity are ligated, and the biotin label is used to selectively enrich for ligated junctions. Finally, isolated ligation products are prepared for high-throughput sequencing, which enables the mapping of pair-wise chromatin interactions genome-wide. Over the past decade, “next-generation” sequencing has become cheaper and easier to perform, enabling more interactions to be sampled to obtain higher resolution in chromatin interaction maps. Here, we provide an in-depth guide to performing an up-to-date Hi-C procedure on mammalian cell lines. These protocols include recent improvements that increase the resolution potential of the assay, namely by enhancing cross-linking and using a restriction enzyme cocktail. These improvements result in a versatile Hi-C procedure that enables the detection of genome folding features at a wide range of distances. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Fixation of nuclear conformation
Basic Protocol 2 : Chromosome conformation capture
Basic Protocol 3 : Hi-C sequencing library preparation
INTRODUCTION
Chromatin is actively folded and organized, enabling an entire genome to fit within the confined volume of a cell. Chromatin organization can influence biological processes. For example, the spatial organization of chromatin can enable specific gene expression patterns and, in multicellular eukaryotes, can dictate cellular identity. Chromosome conformation capture (3C) allows the study of genomic interactions in 3D nuclear space. In this technique, nuclear conformation is chemically fixed prior to enzyme-based DNA digestion and subsequent ligation of fragments in close 3D proximity. A major improvement in conformation capture methodology was the replacement of the PCR readout used in the original 3C method (Dekker, Rippe, Dekker, & Kleckner, 2002) with (whole-genome) sequencing, (Lieberman-Aiden et al., 2009) enabling the capture of all possible pair-wise genomic interactions (Fig. 1). In earlier protocols, Hi-C was performed using restriction enzymes that recognize 6-bp DNA sequences, limiting data resolution to ∼10 kb. Also, to reduce spurious ligation events, digested chromatin was diluted into large volumes before proximity ligation, which was tedious and costly (Belton et al., 2012). Two breakthrough improvements to the Hi-C protocol were (1) the ability to perform proximity ligation in situ within intact nuclei, drastically reducing the number of cells and the reagent volumes needed, and (2) the use of a 4-base cutter, which greatly increased the resolution of the assay (Rao et al., 2014). More recently, the Hi-C protocol has been re-evaluated to comprehensively assess the effects of different fixation methods and restriction enzymes on data quality (Akgol Oksuz et al., 2020). This study led us to further improve the Hi-C protocol by incorporating disuccinimidyl glutarate (DSG) in addition to formaldehyde fixation and employing both DpnII and DdeI restriction enzymes to digest chromatin. The result is a versatile Hi-C protocol that outperforms traditional in situ Hi-C in the detection of loops, topologically associating domains (TADs), and compartments.
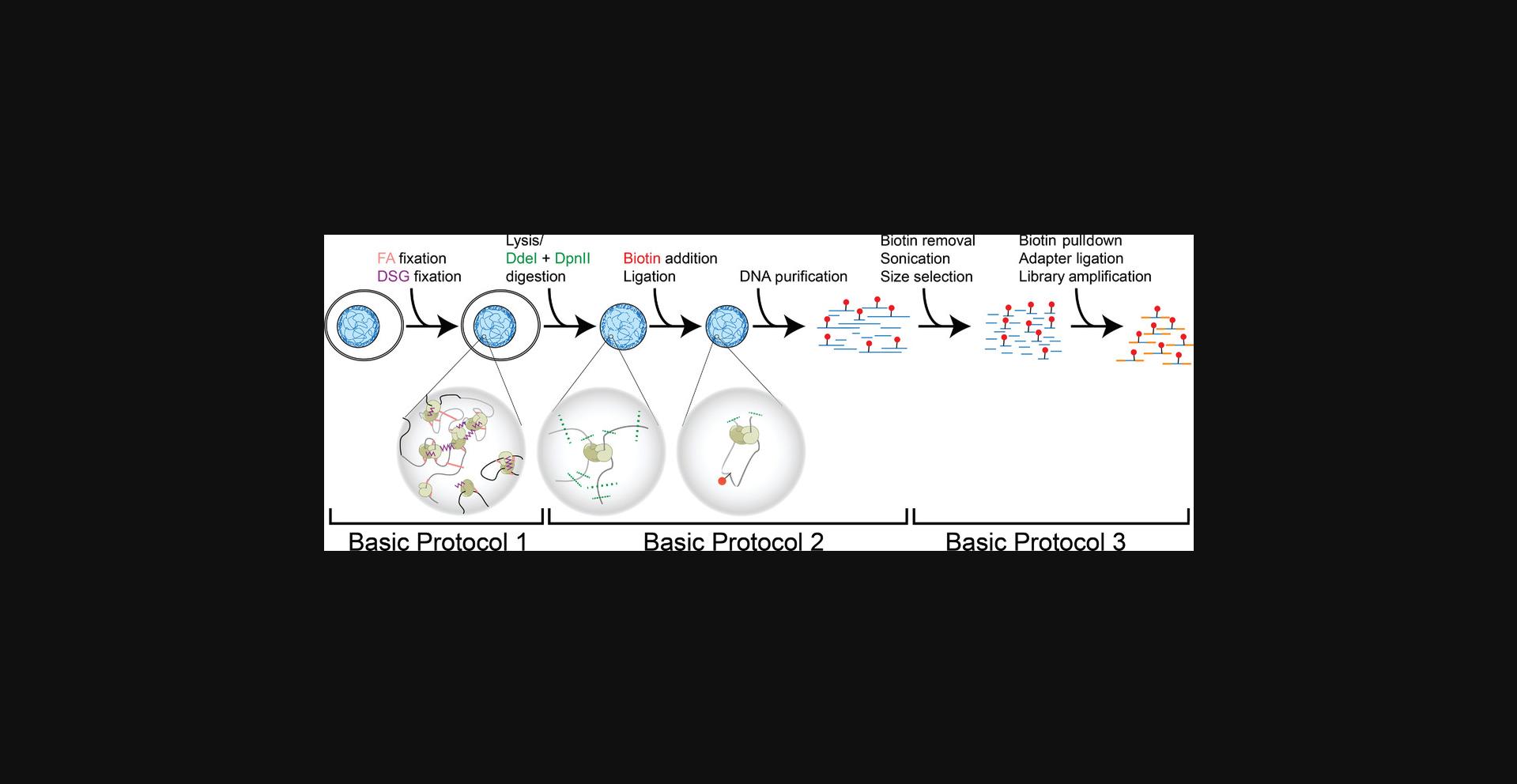
The Hi-C procedure detailed here is divided into three basic protocols (Fig. 1). Basic Protocol 1 details the fixation of chromatin from mammalian cell lines through the use of two different chemical cross-linkers. Chromatin is then restriction digested, and fragments in close 3D proximity are allowed to ligate (Basic Protocol 2). The end of Basic Protocol 2 is marked by the isolation of ligated chromatin and an evaluation of chromatin conformation capture quality. Basic Protocol 3 then describes the synthesis of Illumina sequencing libraries from the proximity-ligated material generated in Basic Protocol 2. This protocol does not, however, cover the downstream analysis of sequenced read pairs, which is outside the scope of this article. Briefly, sequenced DNA pairs can be mapped, filtered, binned, and balanced (iteratively corrected), as outlined by Lajoie, Dekker, and Kaplan (2015). A description of the most up-to-date analyses and tools can be found in Akgol Oksuz et al. (2020).
It is worth mentioning that although the Hi-C assay described here is performed on mammalian cell lines, minor adaptations of previous protocols have allowed Hi-C to be applied in a wide variety of organisms (Belton & Dekker, 2015; Marinov et al., 2021; Moissiard et al., 2012; Nand et al., 2021; Wang et al., 2015).
STRATEGIC PLANNING
The specific chromatin organization features that the user is interested in will drive several aspects of the Hi-C experimental design. If the goal of the experiment is only to assess compartmentalization, the user might consider using a 6-bp-cutting restriction enzyme such as HindIII (Akgol Oksuz et al., 2020), to maximize compartment detection. On the other hand, if the user is interested only in maximizing the resolution of loops, then the micrococcal nuclease chromosome conformation assay (Micro-C; Hsieh et al., 2015; Krietenstein et al., 2020) would be the most appropriate assay. However, if all of these features are of interest, the protocol described herein will strike a balance for the detection of both large- and small-scale features.
Given that the size of a given genome is fixed, the two factors that determine the resolution of a Hi-C heatmap are the number of sequenced reads and the size of the bins that hold them, with a reduction of the bin size requiring more reads to fill them. For example, doubling the resolution of a Hi-C interaction matrix will require 4× (22×) the number of reads, thus quadrupling the sequencing costs. An important requirement for increased resolution is that the enzyme(s) used for genome fragmentation generate fragments that are not, on average, larger than the bin size. For instance, because HindIII digests the human genome into ∼10-kb fragments, individual fragments cannot be assigned to independent 1-kb bins, no matter how many reads are sequenced. Therefore, it is important to tailor the choice of restriction enzymes to the desired resolution.
A second, related consideration is the depth of sequencing required for a particular experiment. As a rule of thumb, more sequencing will increase the resolution of the data. However, this is limited by the complexity of the library, which can be increased by raising the number of input cells for the assay. We prefer preparing replicate libraries in parallel over increasing the number of cells and reaction volumes (i.e., preparing four libraries of 5 million cells versus a single library of 20 million cells). On the other hand, if the priority of the experiment is to maximize the number of samples rather than the sequencing depth, a single lane of an Illumina flow cell can be loaded with multiple, indexed samples. Indexing each sample individually while adhering to Illumina index pooling guides provides the flexibility to either sequence a single sample or pool several samples for sequencing on a single flow cell. Pooling of samples will split the number of reads by the number of samples, generating fewer reads per sample. Typically, this strategy is used to assess the quality of libraries before deciding to sequence more deeply. For more information regarding the relationship between sequencing depth and resolution of various Hi-C structures, see the Understanding Results section.
For Basic Protocol 1, we describe two alternative approaches: one for use on cells that grow in monolayer and one for use on cells that grow in suspension. It should be noted that when cells grow in a monolayer, cell counting immediately before cross-linking can be difficult due to clumping of cells after they are scraped from the plate. Therefore, it is important to know how many cells to seed to attain a subconfluent monolayer of 5 million cells at harvest. We prefer scraping over trypsinization because cell release may affect the nuclear conformation. Although cells in suspension can be grown in a variety of containers, we use tissue culture flasks because they are easy to handle and can accommodate large volumes if needed. Listed under Materials are 175-cm2 flasks, which can hold a maximum density of ∼2 × 107 cells, allowing active growth of 5 × 106 cells.
The entire Hi-C procedure, from fixation to sequencing, can be comfortably completed within 6 days if only a couple of samples are handled simultaneously. With experience, this protocol could be further condensed to 4-5 days by limiting the number of stop points and quality control checkpoints. A well-organized and experienced user can process 16 samples or more in parallel. For more information and tips regarding stop points and timelines, refer to Time Considerations. We present several possible adaptations to our core protocol in regard to a low number of input cells, ultra-deep sequencing, and alternative restriction enzyme(s) to tailor the protocol to the specific question the researcher seeks to answer.
Basic Protocol 1: FIXATION OF NUCLEAR CONFORMATION
This protocol describes the methodology used to capture chromosome conformation before proximity ligation. Cells grown in appropriate medium are cross-linked sequentially with formaldehyde and disuccinimidyl glutarate (DSG). Formaldehyde reacts with amino and imino groups of both protein and DNA, and can efficiently produce cross-links within a 2-Å radius, making it suitable for capturing proximal interactions (reviewed in Hoffman, Frey, Smith, & Auble, 2015). On the other hand, the NHS-esters in DSG react with the primary amines on proteins and can capture amine-amine interactions within 8 Å (Strang, Cushman, DeRubeis, Peterson, & Pfaffinger, 2001). Using the combination of DSG and formaldehyde has been demonstrated to better preserve chromatin contacts and increase data quality at various resolutions when compared to the use of formaldehyde alone (Akgol Oksuz et al., 2020; Hsieh et al., 2015). After cross-linking, cells are typically snap-frozen for storage at −80°C. However, we have noticed that using freshly fixed cells may enhance data quality. This is likely due to the freezing and thawing of samples.
Materials
-
Cells growing in appropriate growth medium
-
HBSS (Gibco, cat. no. 14025-092)
-
DPBS (Gibco, cat. no. 14190-144)
-
37% formaldehyde (Fisher, cat. no. BP531-500)
-
2.5 M glycine in Milli-Q water (Sigma, cat. no. G8898-1KG)
-
DSG (ThermoScientific, cat. no. 20593)
-
DMSO (Sigma, cat. no. D2650-5 × 10ML)
-
0.05% (w/v) BSA in DPBS
-
150 × 25-mm culture plates (Corning, cat. no. 430599; for monolayer cells) or 175-cm2 culture flask (Falcon, cat. no. 353112; for suspension cells)
-
CO2 incubator (Cell Culture)
-
50-ml conical tubes
-
Ice bucket
-
Cell scraper (Falcon, cat. no. 353089)
-
Tabletop centrifuge
-
Rotator (Argos Technologies, cat. no. EW-04397-40) or rocking platform
-
Hemocytometer
-
1.7-ml microcentrifuge tubes (Axygen, MCT-175-C)
-
Liquid nitrogen
Starting from cells in monolayer
Cross-linking with formaldehyde
1a. The day before the experiment, seed cells in 150-mm plates in appropriate medium to have 5 × 106 cells per plate for harvest the next day, and incubate overnight.
2a. Aspirate medium from the plate and wash twice with ∼10 ml HBSS.
3a. Immediately before cross-linking, prepare a 1% formaldehyde cross-linking solution in a 50-ml tube by combining 22.5 ml HBSS and 625 µl of 37% formaldehyde. Mix well.
4a. Pour the 1% formaldehyde solution (∼23.125 ml) into each 150-mm plate.
5a. Incubate exactly 10 min at room temperature, gently rocking the plates by hand every 2 min.
6a. To quench the cross-linking reaction, add 1.25 ml of 2.5 M glycine (to 128 mM final) and mix well by gently swirling the plate.
7a. Incubate the plates 5 min at room temperature, and then place on ice for at least 15 min to stop the cross-linking completely.
8a. Scrape the cells from the plates with a cell scraper.
9a. Using a pipet, transfer the cell suspension to a 50-ml conical tube and centrifuge 10 min at 1000 × g , room temperature.
10a. Wash the cell pellet once with 10 ml DPBS, pipetting to resuspend, and centrifuge 10 min at 1000 × g , room temperature. Aspirate and discard the supernatant. Proceed immediately to DSG cross-linking (step 11).
Starting from cells in suspension
Cross-linking with formaldehyde
1b. The day before the experiment, seed enough cells in a 175-cm2 flask in appropriate medium to have 5 × 106 cells per flask at harvest, and incubate overnight.
2b. Immediately before harvest, count cells with a hemocytometer and transfer 5 × 106 cells to a 50-ml conical tube.
3b. Gently pellet the cells by centrifuging 10 min at 300 × g , room temperature.
4b. Prepare a 1% formaldehyde cross-linking solution by quickly adding 1.25 ml of 37% formaldehyde to 45 ml HBSS, and immediately mix by inverting the tube several times.
5b. Resuspend the cell pellet completely using the 1% formaldehyde cross-linking solution prepared in step 4b (∼46.25 ml) by pipetting up and down.
6b. Incubate exactly 10 min at room temperature on a rotator.
7b. Quench the cross-linking reaction by adding 2.5 ml of 2.5 M glycine (final concentration of 128 mM) in one shot. Mix well by inverting the tube.
8b. Incubate 5 min at room temperature and then incubate at least 15 min on ice to stop the cross-linking completely.
9b. Centrifuge the cross-linked cells 10 min at 1000 × g , room temperature. Discard the supernatant by aspiration.
10b. Wash pellet once with 5-10 ml DPBS and centrifuge cells 10 min at 1000 × g , room temperature. Discard the supernatant completely. Proceed immediately to DSG cross-linking (step 11).
Cross-linking with disuccinimidyl glutarate (DSG)
11.Resuspend the cell pellet in 9.9 ml DPBS. Add 100 µl DSG 300 mM (to 3 mM final) and mix by inversion.
12.Incubate samples 40 min at room temperature on a rotator.
13.Add 1.925 ml of 2.5 M glycine (to 400 mM final), mix well by inversion, and incubate 5 min at room temperature.
14.Centrifuge cells 15 min at 2000 × g , room temperature, and remove supernatant.
15.Resuspend the pellet in 1 ml of 0.05% BSA-DPBS and transfer to a 1.7-ml tube.
16.Centrifuge cells 15 min at 2000 × g , 4°C, and then remove supernatant completely.
17.Snap-freeze in liquid nitrogen, and either store the pellet at −80°C or continue immediately to Basic Protocol 2.
Basic Protocol 2: CHROMOSOME CONFORMATON CAPTURE
This protocol describes the steps that follow cross-linking of cells up until DNA purification of proximity-ligated chromatin fragments. Cross-linked cells obtained in Basic Protocol 1 are first lysed and mechanically disrupted before being subjected to restriction enzyme digestion. The use of both DpnII and DdeI restriction enzymes yields a fragment length distribution that allows high-quality data analysis of genomic structures, ranging from loops to chromosome domains. The filled-in overhangs are then ligated to fragments that are in close 3D proximity. After reversal of cross-linking, DNA is purified using a protocol adapted from Moore & Dowhan (2002) and the resulting sample is treated with RNase. Quality control of the conformation capture process marks the final step of this protocol. Here, the quality of the input chromatin is assessed, as well as the extent to which chromatin is restriction-digested and ligated. Using gel electrophoresis, both controls are compared to the proximity-ligated material (Hi-C sample). Purified proximity-ligated samples can then be stored at −20°C before continuing with Basic Protocol 3.
Materials
-
Cell pellets (Basic Protocol 1)
-
Lysis buffer (see recipe)
-
10× NEBuffer 3.1 (New England Biolabs, cat. no. B7203S), or other appropriate buffer (see Critical Parameters)
-
1% (w/v) SDS, diluted from 20% (Fisher, cat. no. BP13111)
-
10% Triton X-100 (Sigma, cat. no. 93443)
-
DdeI (New England Biolabs, cat. no. R0175L)
-
DpnII (New England Biolabs, cat. no. R0543M)
-
10 mM dCTP (Invitrogen, cat. no. 56173)
-
10 mM dGTP (Invitrogen, cat. no. 56174)
-
10 mM dTTP (Invitrogen, cat. no. 56175)
-
0.4 mM biotin-14-dATP (Invitrogen, cat. no. 19524-016)
-
5 U/µl Klenow DNA polymerase (New England Biolabs, cat. no. M0210L)
-
5× ligation buffer (Invitrogen, cat. no. Y90001)
-
10 mg/ml BSA (New England Biolabs, cat. no. B9000S)
-
Milli-Q water
-
1 U/µl T4 DNA ligase (Invitrogen, cat. no. 15224090)
-
TLE (see recipe)
-
10 mg/ml proteinase K in Milli-Q water (Invitrogen, cat. no. 25530-031)
-
Phenol/chloroform/isoamyl alcohol (Invitrogen, cat. no. 15593-049)
-
3 M sodium acetate, pH 5.2
-
100% ethanol (Fisher, cat. no. 04355222)
-
10 mg/ml RNase A, DNase and protease free (Thermo Scientific, cat. no. EN0531)
-
Agarose
-
TBE (see recipe)
-
1-kb ladder (New England Biolabs, cat. no. N3232L)
-
6× Orange G gel loading dye (see recipe)
-
Dounce homogenizer (DWK Life Sciences, cat. no. 8853010002/8853030002)
-
1.7-ml microcentrifuge tubes (Axygen, MCT-175-C)
-
Tabletop centrifuge
-
Vortex
-
Thermomixer (Eppendorf, cat. no. 5382000023)
-
15-ml conical tube (Denville, cat. no. C1017-P)
-
15- and 2-ml phase-lock tube (Qiagen, cat. nos. 129065 and 129056)
-
35-ml Beckman ultracentrifuge tube (Beckman Coulter, cat. no. 357002)
-
Dry ice
-
Ultracentrifuge
-
Amicon Ultra-0.5 Centrifugal Filter Unit (EMD Millipore, cat. no. UFC500396)
-
Refrigerated microcentrifuge
-
Agarose gel electrophoresis apparatus
-
Densitometer, Qubit, or Nanodrop apparatus
Cell lysis and chromatin digestion
1.Resuspend the cross-linked cell aliquots from Basic Protocol 1 (step 17) by pipetting in 1 ml of ice-cold lysis buffer containing 10 µl protease inhibitor cocktail. Transfer the mix to a dounce homogenizer and incubate 15 min on ice.
2.Dounce homogenize the cells on ice with pestle A (the large-clearance pestle). Slowly move the pestle up and down 30 times, incubate on ice 1 min to allow cells to cool down, and then continue with another 30 strokes.
3.Transfer the lysate to a 1.7-ml microcentrifuge tube.
4.Centrifuge the lysed suspension 5 min at 2500 × g , room temperature.
5.Discard the supernatant and resuspend the wet pellet by vortexing.
6.Resuspended the pellet in 500 µl ice-cold 1× NEBuffer 3.1 and centrifuge 5 min at 2500 × g , room temperature.
7.Repeat step 6 for a second wash.
8.Resuspend pellet in 1× NEBuffer 3.1 to a final volume of 360 µl by pipetting.
9.Save 18 µl of each lysate for “CHROMATIN INTEGRITY CONTROL” (CI) and store these at 4°C.
10.Add 38 µl of 1% SDS to each Hi-C sample (380 µl total sample volume) and mix carefully by pipetting to avoid making bubbles.
11.Incubate samples in Thermomixer for exactly 10 min at 65°C without shaking, to open up chromatin.
12.Immediately place tubes on ice and add 43 µl of 10% Triton X-100 to each Hi-C sample (423 µl total sample volume) to quench the SDS. Gently mix by pipetting, avoiding making bubbles.
13.Add 40 µl of 10 U/µl DdeI, 8 µl of 50 U/µl DpnII, and 4 µl of 1× NEBuffer 3.1 (475 µl total sample volume) and carefully mix by pipetting.
14.Digest the chromatin overnight (∼16 hr) at 37°C in a Thermomixer with interval shaking (900 rpm, 30 s on, 4 min off).
Biotinylation of DNA ends
15.After the overnight restriction enzyme digestion is complete, incubate the Hi-C samples 20 min at 65°C to deactivate the endonuclease enzymes.
16.During the incubation, prepare the biotin fill-in master mix as described below (amount per reaction given):
| Reagent | Amount |
|---|---|
| 10× NEBuffer 3.1 | 7 µl |
| 10 mM dCTP | 1.5 µl |
| 10 mM dGTP | 1.5 µl |
| 10 mM dTTP | 1.5 µl |
| 0.4 mM biotin-14-dATP* | 37.5 µl |
| Milli-Q water | 11 µl |
| Klenow DNA polymerase | 10 µl |
17.After incubation (step 15), place the Hi-C samples immediately on ice.
18.Transfer 10 µl from each Hi-C sample to a 1.7-ml microcentrifuge tube, label it as the “DIGESTION CONTROL” (DC) for that sample, and store at 4°C.
19.Briefly spin down the tubes to remove condensation from the lid. Add 70 µl of the biotin fill-in mix (step 16) to each Hi-C sample (535 µl total sample volume), and mix gently by pipetting without making bubbles.
20.Incubate the Hi-C samples 4 hr at 23°C in a Thermomixer (900 rpm, 30 s on, 4 min off).
Ligation of proximal DNA fragments
21.During the biotin fill-in incubation (step 20), prepare the ligation mix as described below (amount per reaction given):
| Reagent | Amount |
|---|---|
| 5× ligation buffer | 240 µl |
| 10% Triton X-100 | 120 µl |
| 10 mg/ml BSA | 12 µl |
| Milli-Q water | 243 µl |
| 1 U/µl T4 DNA ligase | 50 µl |
22.Add 665 µl of ligation mix to each Hi-C sample (to 1200 µl total sample volume) and mix gently by pipetting.
23.Incubate the Hi-C samples 4 hr at 16°C in a Thermomixer with interval shaking (900 rpm, 30 s on, 4 min off).
Reversal of cross-linking
24.Bring the volumes of both the CI and DC samples (stored at 4°C in steps 9 and 18, respectively) to 50 µl each with TLE.
25.To each CI and DC sample, add 10 µl of 10 mg/ml proteinase K and incubate overnight at 65°C with interval shaking (900 rpm, 30 s on, 4 min off). Then keep at room temperature.
26.To each Hi-C sample (from step 23), add 50 µl of 10 mg/ml proteinase K and incubate 2 hr at 65°C with interval shaking (900 rpm, 30 s on, 4 min off).
27.Add another 50 µl of 10 mg/ml proteinase K to each Hi-C sample and continue incubating overnight at 65°C.
DNA purification
28.Remove the Hi-C samples from 65°C and allow tubes to reach room temperature.
29.Transfer each sample to an individual 15-ml conical tube and add 2 vol (2.6 ml) phenol/chloroform/isoamyl alcohol to each.
30.Vortex each Hi-C sample for 1 min and then slowly pour contents into a 15-ml phase-lock tube.
31.Centrifuge the Hi-C samples in 15-ml tubes for 5 min at 1500-3500 × g in a benchtop centrifuge.
32.Carefully pour the aqueous phase from each Hi-C sample into a 35-ml Beckman round-bottom ultracentrifuge tube and bring to a total volume of 1250 µl with Milli-Q water.
33.Add 1/10 vol (125 µl) 3 M sodium acetate to each sample and mix well by inversion.
34.Add 2.5 vol (3.4 ml) ice-cold 100% ethanol and mix well by inversion.
35.Incubate the Hi-C samples ∼15 min on dry ice (to avoid sample solidification).
36.Centrifuge 30 min at 18,000 × g , 4°C.
37.Remove supernatant completely and discard.
38.Air-dry the Hi-C samples for ∼10 min or until visibly dry.
39.Solubilize each pellet in 450 µl TLE and transfer to an Amicon Ultra-0.5 Centrifugal Filter Unit.
40.Centrifuge Amicon columns 10 min at 14,000 × g (maximum speed) in a microcentrifuge and discard flowthrough.
41.Flush each Beckman tube with another 450 µl TLE and transfer to its respective Amicon column for washing.
42.Centrifuge Amicon columns 10 min at 14,000 × g (maximum speed) in a microcentrifuge and discard flowthrough.
43.Wash each column with 450 µl TLE, centrifuging at 14,000 × g (maximum speed) and discarding the flowthrough.
44.Add 80 µl TLE to the column, flip column into a fresh collection tube, and centrifuge 2 min at 14,000 × g (maximum speed) in a microcentrifuge.
45.Add 1 µl of 1 mg/ml RNase A (a 10-fold dilution of 10 mg/ml stock) to each Hi-C sample and incubate at least 20 min at 37°C on a heat block, water bath, or Thermomixer.
46.Remove the samples from 37°C and store at 4°C until needed for quality control step.
Quality control of chromatin integrity, digestion, and proximity ligation
47.After cross-link reversal (see step 25), cool CI and DC samples to room temperature and transfer to pre-spun 2-ml phase-lock tubes.
48.Add 200 µl phenol/chloroform/isoamyl alcohol and mix control samples by vortexing for 1 min.
49.Centrifuge tubes 5 min at 14,000 × g (maximum speed) in microcentrifuge.
50.Transfer the aqueous phase from each sample (∼50 µl) to a fresh 1.7-ml microcentrifuge tube.
51.Add 1 µl of 1 mg/ml RNase A and incubate at least 30 min at 37°C.
52.Load the control samples, Hi-C samples (from step 46), and 1-kb ladder, each with 6× Orange G gel loading dye added to 1× concentration, into wells of a 0.8% agarose/TBE gel. Run the gel in 0.5× TBE buffer using an agarose gel electrophoresis apparatus.
| Sample | Contents |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
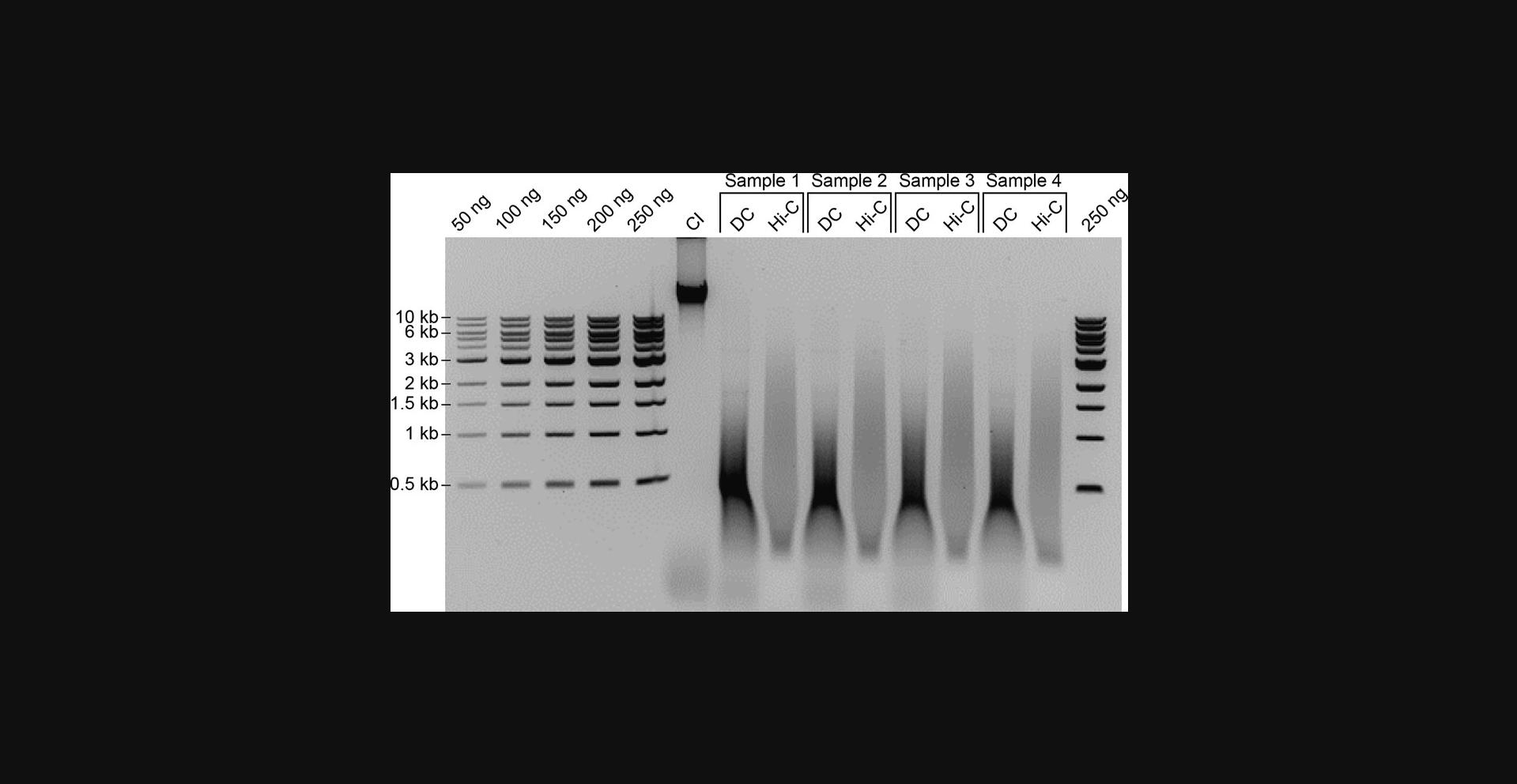
53.Quantify DNA by densitometry from the gel directly or by using other methods such as Qubit or Nanodrop.
Basic Protocol 3: Hi-C SEQUENCING LIBRARY PREPARATION
This protocol describes how to transform the ligation products generated in Basic Protocol 2 into a format suitable for Illumina sequencing. Before ligation junctions can be selected, biotin must be removed from unligated ends (see discussion of partial digests in Background Information). Because Illumina sequencing platforms require libraries to have a specific fragment size range for optimal cluster formation, Hi-C samples must be fragmented by sonication and further size-selected using AMPure XP beads. DNA damage induced by sonication is then repaired, and sequencing adapters are ligated to freshly A-tailed ends. We provide here methodology for the generation and ligation of indexed Illumina TruSeq adapters. We have also successfully generated Hi-C libraries through ligation of Illumina PE adapters, and should note that other methods could potentially be used to ligate adapters compatible with Illumina sequencing. Once adapters are ligated, a PCR titration is performed to assess the optimal number of cycles needed to attain enough material for sequencing while minimizing PCR duplicates. Final library amplification is performed using this optimized number of PCR cycles. Samples are then purified, and primer dimers are removed using AMPure XP beads, yielding a library that is ready for sequencing.
Materials
- Hi-C DNA samples from Basic Protocol 2 (step 46)
- 10× NEBuffer 2.1 (New England Biolabs, cat. no. B7002S)
- 1 mM dATP (Invitrogen, cat. no. 56172)
- 1 mM dGTP (Invitrogen, cat. no. 56174)
- T4 DNA polymerase (New England Biolabs, cat. no. M0203L)
- Milli-Q water
- Agencourt AMPure XP beads, 60 ml (Beckman Coulter, A63881)
- TLE (see recipe)
- 70% (v/v) ethanol, freshly prepared
- Low-molecular-weight DNA ladder (New England Biolabs, cat. no. N3233L)
- Agarose
- TBE (see recipe)
- Ethidium bromide (Fisher, cat. no. BP1302-10)
- 6× Orange G gel loading dye (see recipe)
- 10× ligation buffer (New England Biolabs, cat. no. B0202S)
- 25 mM dNTP mix (Invitrogen, cat. no. 10297117)
- 3 U/µl T4 DNA polymerase (New England Biolabs, cat. no. M0203L)
- 10 U/µl T4 polynucleotide kinase (New England Biolabs, cat. no. M0201L)
- 5 U/µl Klenow DNA polymerase (New England Biolabs, cat. no. M0210L)
- MyOne Streptavidin C1 beads (Invitrogen, cat. no. 65001)
- Tween wash buffer (TWB; see recipe)
- 2× binding buffer (BB; see recipe)
- 5 U/µl Klenow fragment (3→5′ exo–; New England Biolabs, cat. no. M0212L)
- 5× T4 ligation buffer (Invitrogen, cat. no. Y90001)
- TruSeq adapter oligos (Table 2)
- 1 U/µl T4 DNA ligase (Invitrogen, cat. no. 15224090)
- 5× annealing buffer (see recipe)
- Pfu Ultra II Fusion HotStart DNA Polymerase and 10× Pfu Ultra II Buffer (Agilent, cat. no. 600674)
| Oligo | Sequencea |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- a
/5PHOS/, 5′ phosphate; *, phosphorothioate bond.
- PCR tubes
- PCR thermocycler
- 130-µl Covaris sonication tubes (micro tubes AFA fiber with snap-cap; Covaris, 520045/520077)
- Covaris sonicator (Covaris, E220 evolution/M220)
- 1.7-ml LoBind tubes (Eppendorf, cat. no. 22431021)
- Vortex
- Rotator (Argos Technologies, cat. no. EW-04397-40)
- Magnetic particle separator (ThermoFisher, cat. no. 12321D)
- Agarose gel electrophoresis apparatus
- PCR strip tubes (Bio-Rad, cat. no. TBS0201/TCS0803)
Removal of biotin from unligated ends
1.Prepare biotin removal reactions in PCR tubes as described below:
| Reagent | Amount |
|---|---|
| Hi-C DNA sample | 5-15 µg |
| 10× NEBuffer 2.1 | 13 µl |
| 1 mM dATP | 3.25 µl |
| 1 mM dGTP | 3.25 µl |
| 3000 U/ml T4 DNA polymerase | 13 µl |
| Milli-Q water | to 130 µl |
2.Distribute two 65-µl aliquots from each 130-µl reaction across two PCR tubes.
3.Transfer to a thermocycler and subject each reaction to the following cycling parameters:
| Time | Temperature |
|---|---|
| 4 hr | 20°C |
| 20 min | 75°C |
| Hold | 4°C |
4.Proceed immediately to sonication, or store samples at 4°C (days to weeks) or −20°C (long term).
Sonication
5.Pool the split samples from step 4 (130 µl total sample volume) and transfer to a Covaris sonication tube.
6.Sonicate samples on either on a Covaris E220 evolution or a Covaris M220 using the following parameters, to achieve a fragment size distribution <500 bp:
| Parameter | Covaris M220 | Covaris E220 evolution |
|---|---|---|
| Peak incident power (W) | 50 W | 175 W |
| Duty cycle/duty factor | 20% | 10% |
| Intensity | – | 5 |
| Cycles per burst | 200 | 200 |
| Treatment time (s) | 180 | 180 |
Size selection
7.Transfer sonicated samples from the Covaris tubes into a 1.7-ml LoBind microcentrifuge tube and label them UF (“Upper Fraction”).
8.Bring each sample to a total volume of 500 µl with TLE.
9.Add 400 µl of AMPure XP mixture to each tube to attain a 0.8× ratio of AMPure volume to sample volume.
10.Mix tubes by vortexing and incubate 10 min at room temperature on the rotator.
11.Place the tubes on a magnetic particle separator (MPS) for 5 min at room temperature.
12.During this incubation, prepare fresh 1.7-µl tubes for each sample. Add 500 µl AMPure beads to each one and label them LF (“Lower Fraction”).
13.Incubate tubes from step 12 on MPS for 5 min.
14.Discard the supernatant from the tubes in step 13 and resuspend the captured beads in 150 µl AMPure XP beads.
15.Collect the supernatant from step 11 (<300 bp) and transfer to the corresponding tube prepared to select the Lower Fraction (step 14).
16.Vortex the Lower Fraction tubes to mix and incubate 10 min at room temperature on the rotator.
17.After incubation, place the Lower Fraction tubes on the MPS for 5 min at room temperature.
18.Discard the supernatant from the Lower Fraction tubes and spin tubes down briefly using a microcentrifuge.
19.Wash the beads from both Upper and Lower Fractions twice with 200 µl of 70% ethanol, capturing beads against the MPS for 5 min each time.
20.Completely remove ethanol and air dry the beads on the MPS.
21.Resuspend both fractions in 50 µl TLE buffer and incubate 10 min at room temperature on the bench, flicking to mix every 2 min to elute the DNA.
22.Capture beads from both fractions on the MPS for 5 min.
23.Transfer supernatant from each sample to a fresh 1.7-ml LoBind tube.
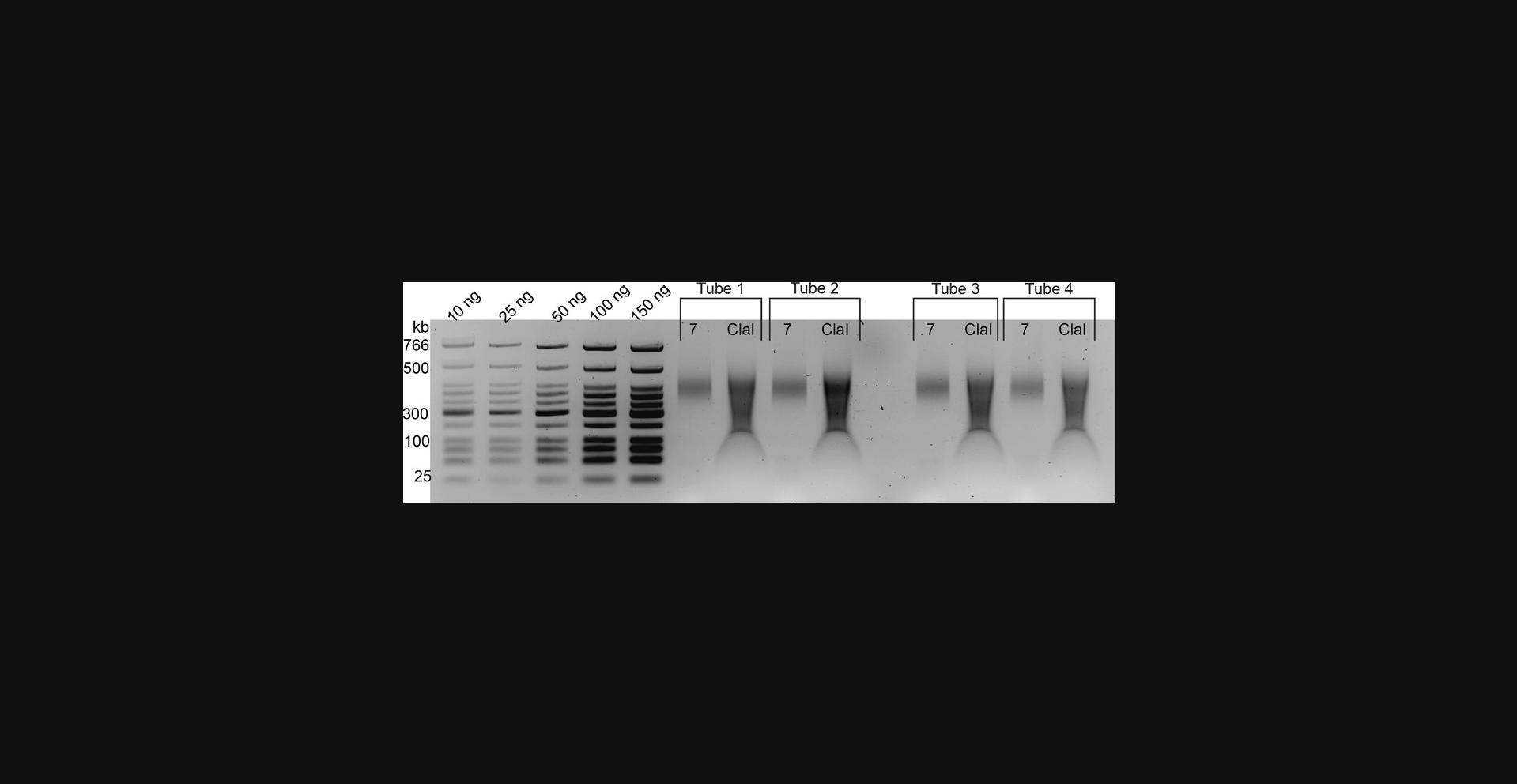
24.Run samples on a 2% agarose/TBE gel as described below to assess the quality and to quantify size-selected samples. A typical gel can be found in Figure 3.
| Sample |
|
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
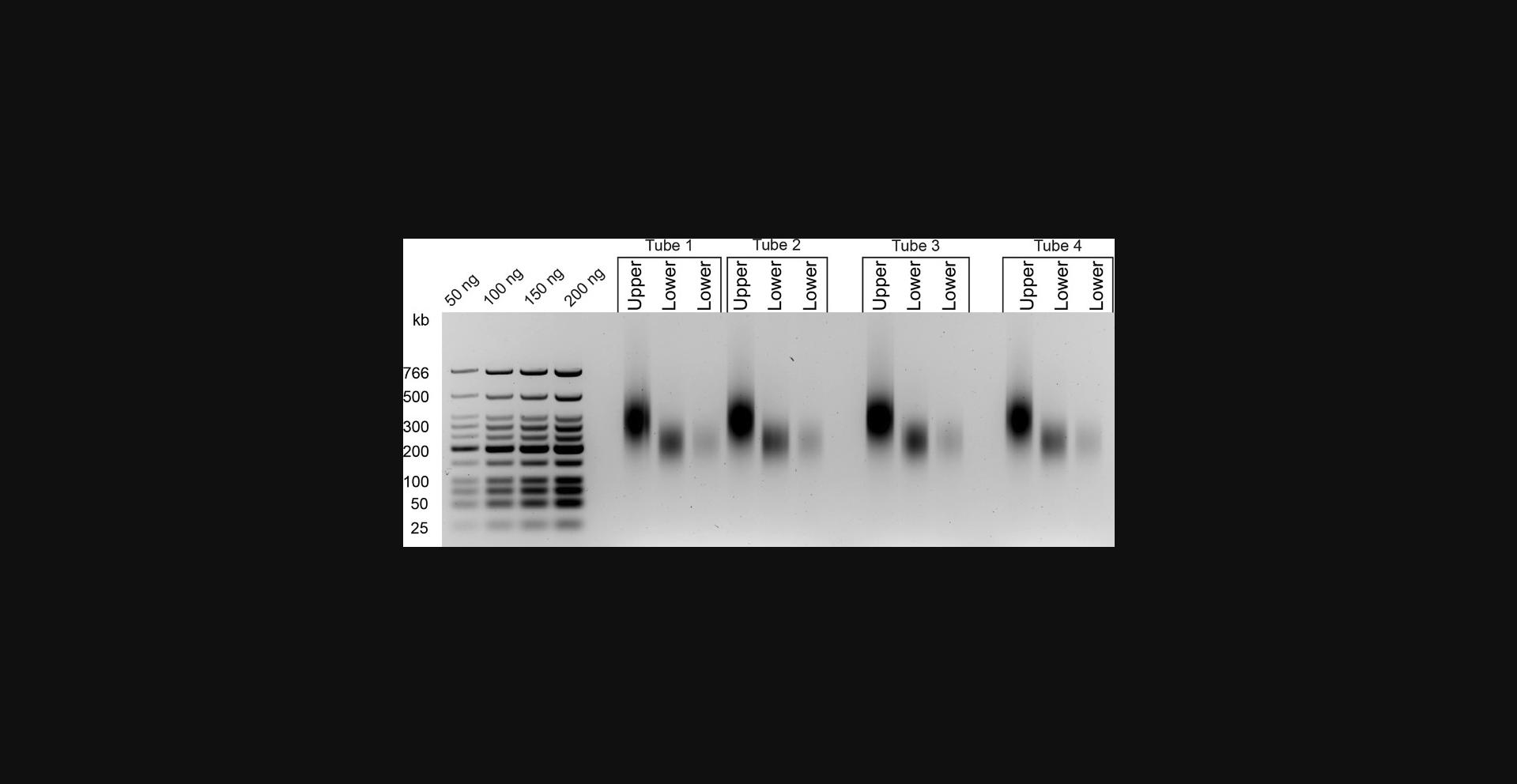
25.Quantify DNA from the gel directly using a standard curve from the known DNA ladder input, or use other methods such as Qubit or Nanodrop.
End repair
26.Prepare the end-repair mix as described below (amount per reaction given):
| Reagent | Amount |
|---|---|
| 10× ligation buffer | 7 µl |
| 2.5 mM dNTP mix | 7 µl |
| 3 U/µl T4 DNA polymerase | 2.5 µl |
| 10 U/µl T4 polynucleotide kinase | 2.5 µl |
| 5 U/µl Klenow DNA polymerase | 0.5 µl |
| Milli-Q water | 4.5 µl |
27.Transfer the remaining 46 µl of size-selected Hi-C libraries (Lower Fraction) from step 23 to PCR strip tubes and add 24 µl of end repair mix. Incubate in a thermocycler as described below:
| Time | Temperature |
|---|---|
| 30 min | 20°C |
| 20 min | 75°C |
| Hold | 4°C. |
28.Once the program is completed, place samples on ice until pulldown.
Pulldown of biotinylated ligation products
29.Having quantified each Hi-C library after size selection (step 25), determine the amount of MyOne streptavidin beads needed for the library pulldown.
30.Vortex the MyOne Streptavidin C1 beads and transfer the volume of beads calculated for each library in step 29 to individual 1.7-ml LoBind tubes.
31.Wash the beads with 400 µl Tween wash buffer (TWB) by pipetting, and incubate 3 min at room temperature on a rotator.
32.Capture the beads against the MPS for 1 min and discard the supernatant.
33.Resuspend the beads in 400 µl TWB.
34.Capture the beads against the MPS for 1 min and discard the supernatant.
35.Resuspend the beads in 400 µl of 2× binding buffer (BB). Add 330 µl TLE and 70 µl of end-repair reaction containing DNA (from step 28).
36.Incubate the samples 15 min at room temperature on a rotator.
37.Capture the beads against the MPS for 1 min and discard the supernatant.
38.Resuspend the beads in 400 µl of 1× BB.
39.Capture the beads against the MPS for 1 min and discard the supernatant.
40.Wash the beads with 100 µl TLE and transfer to a fresh 1.7-ml LoBind tube.
41.Capture the beads against the MPS for 1 min and discard the supernatant.
42.Resuspend the beads in 41 µl TLE.
A-tailing
43.Prepare the A-tailing mix as described below (amount per reaction given):
| Reagent | Amount |
|---|---|
| Hi-C library on beads (from step 42) | 41 µl |
| 10× NEBuffer 2.1 | 5 µl |
| 10 mM dATP | 1 µl |
| 5 U/µl Klenow fragment (3→5′ exo–) | 3 µl |
44.Transfer reactions to PCR strip tubes and incubate on thermocycler as described below:
| Time | Temperature |
|---|---|
| 30 min | 37°C |
| 20 min | 65°C |
| Hold | 4°C |
45.Immediately after removal from thermocycler, place the PCR tubes on ice and transfer the samples to 1.7-ml LoBind tubes.
46.Capture the beads against the MPS for 1 min and discard the supernatant.
47.Resuspend the beads in 400 µl of 1× ligation buffer, diluted from Invitrogen 5× T4 DNA ligase buffer.
48.Capture the beads against the MPS for 1 min and then discard the supernatant.
49.Resuspend the beads in a final volume of 40 µl of 1× ligation buffer, diluted from Invitrogen 5× T4 DNA ligase buffer.
Annealing of adapter oligos
50.Prepare 100 µM stocks of adapter oligos (Table 2)
51.Combine components of the adapter annealing reaction in PCR strip tubes as described below (amount per reaction given):
| Reagent | Amount |
|---|---|
| TruSeq Adapter-Universal oligo | 20 µl |
| TruSeq Adapter-Indexed oligo | 20 µl |
| 5× annealing buffer | 10 µl |
52.Place adapter annealing reactions in a PCR thermocycler and ramp up temperature at 0.5°C/s to 97.5°C.
53.Hold the temperature at 97.5°C for 150 s.
54.Ramp down temperature at 0.1°C/s for 775 cycles (to ∼20°C).
55.Hold at 4°C afterwards.
56.Dilute the annealed adapters to 15 µM by adding 83 µl of 1× annealing buffer.
57.Store adapters long term at −20°C.
Sequencing adaptor ligation
58.Prepare and combine components of the adapter ligation mix in a 1.7-ml LoBind tube as described below (amount per reaction given):
| Reagent | Amount |
|---|---|
| Hi-C library on beads (from step 49) | 40 µl |
| 5× T4 ligation buffer (Invitrogen, cat. no. Y90001) | 2 µl |
| TruSeq adapter oligos | 5 µl |
| 1 U/µl T4 DNA ligase | 3 µl |
59.Incubate ligation reactions 2 hr at room temperature.
60.Capture the beads against the MPS for 1 min and then discard the supernatant.
61.Wash beads twice with 400 µl TWB.
62.Capture the beads against the MPS for 1 min and discard the supernatant.
63.Resuspend the beads in 200 µl of 1× BB.
64.Capture the beads against the MPS for 1 min and then discard the supernatant.
65.Wash the beads once with 200 µl of 1× NEBuffer 2.1 and transfer to a fresh 1.7-ml LoBind tube.
66.Capture the beads against the MPS for 1 min and discard the supernatant.
67.Resuspend the beads in 20 µl of 1× NEBuffer 2.1.
68.Keep tubes on ice while in use or store at 4°C.
PCR cycle number optimization (titration PCR)
69.Set up 30 µl master mix reactions per sample as described below (amount per reaction given; scale up accordingly):
| Reagent | Amount |
|---|---|
| 10 µM TruSeq-PE1.0 | 0.3 µl |
| 10 µM TruSeq-PE2.0 | 0.3 µl |
| Hi-C library on beads (from step 67) | 1.5 µl |
| 25 mM dNTP mix | 0.3 µl |
| 10× Pfu Ultra II Buffer | 3 µl |
| Pfu Ultra II HiFi DNA Polymerase | 0.5 µl |
| Milli-Q water | 24.1 µl |
70.Distribute 5-µl aliquots of the master mix across four PCR tubes (each will be subjected to a different number of PCR cycles). We usually test 5, 7, 9, and 11 cycles, which should be enough to detect product on gel.
71.Subject each sample replicate to the PCR program outlined below:
|
|
|
|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 min | 72°C |
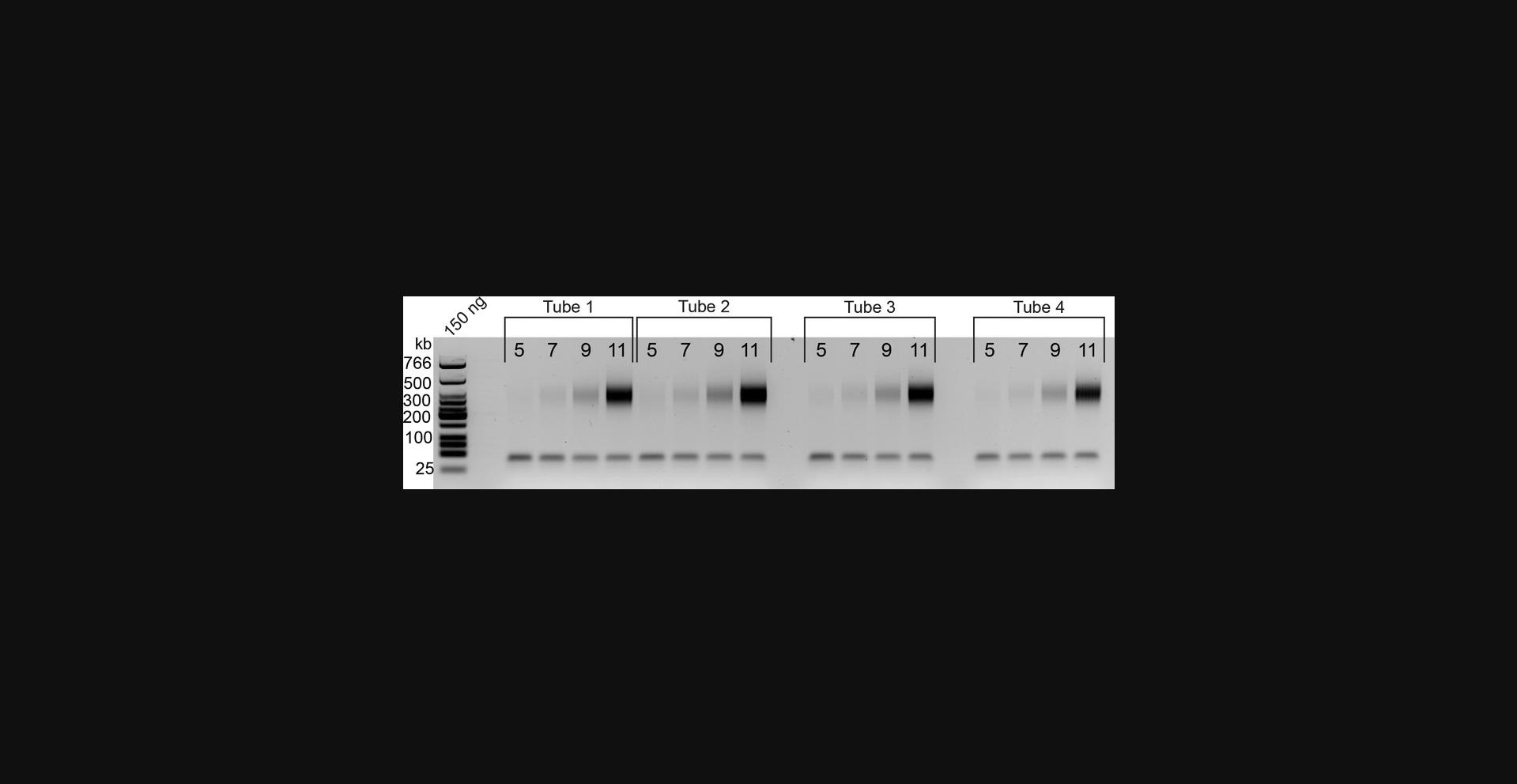
72.Combine each 5 µl sample with 5 µl water and 2 µl of 6× Orange G dye and run on a 2% agarose gel along with 1-3 µl of low-molecular-weight ladder (50 ng/µl). Typical gel results are shown in Figure 4.
Final library PCR amplification
73.After the titration PCR (step 69), ∼18 µl of “Hi-C library on beads” will remain. This allows an additional 12 PCR reactions (360 µl total) under the optimal PCR reaction conditions identified. To amplify the final library for sequencing, set up a reaction as outlined below:
|
|
|---|---|
|
|
|
|
|
|
|
3.6 µl |
|
36 µl |
|
6 µl |
|
289.2 µl |
74.Aliquot the 360-µl mixture from each library into separate PCR tubes/wells and incubate the reactions as in step 71 using the number of cycles determined after PCR titration (step 72.).
75.Once PCR is complete, pool replicate samples into a single 1.7-ml LoBind microcentrifuge tube.
76.Capture beads on the MPS and transfer supernatant to afresh 1.7-ml LoBind microcentrifuge tube.
77.Resuspend the beads in 20 µl of 1× NEBuffer 2.1.
Library cleanup with AMPure beads
78.Dilute the supernatant from step 76 to exactly 360 µl with Milli-Q water.
79.Add 360 µl Agencourt AMPure XP beads per sample (1.0×) and mix well by pipetting.
80.Incubate samples 10 min at room temperature on rotator.
81.Place samples on MPS for 3-5 min at room temperature.
82.Discard the supernatant and wash the beads twice with 200 µl of 70% ethanol, capturing beads against the MPS for 5 min each time.
83.Completely remove ethanol and air dry the beads on the MPS until alcohol is completely evaporated and the pellet looks less shiny (up to 10 min).
84.Resuspend the beads in 30 µl Milli-Q water and incubate 10 min at room temperature on bench, flicking to mix every 2 min, to elute the DNA.
85.Collect the beads on the MPS for 5 min.
86.Transfer supernatant from each sample to a fresh 1.7-ml tube.
87.Run 1 µl of each library on a 2% agarose gel for quantification and to assess fragment size distribution (Fig. 5).
88.Store final libraries at 4°C for a few days or at −20°C long term, or dilute immediately for sequencing submission.
REAGENTS AND SOLUTIONS
Annealing buffer, 5×
- 50 mM Tris·Cl, pH 7.5
- 50 mM NaCl
- 0.5 mM EDTA
- Store at room temperature; stable for years when properly used and tightly closed.
Binding buffer (BB), 2×
- 10 mM Tris·Cl, pH 8.0
- 1 mM EDTA
- 2 M NaCl
- Store at room temperature; stable for years when properly used and tightly closed.
Lysis buffer
- 10 mM Tris·Cl, pH 8.0
- 10 mM NaCl
- 0.2% Igepal CA-630 (MP Biomedicals, cat. no. 198596)
- 1× protease inhibitor cocktail (ThermoFisher, cat. no. 78440)
- Store at 4°C (minus protease inhibitor); stable for months when properly used and tightly closed.
Add protease inhibitor immediately before use only.
Orange G gel loading dye, 6×
- 10 mM Tris·Cl, pH 7.6
- 0.15% Orange G
- 60 ml glycerol
- Store at room temperature; stable for years when properly used and tightly closed.
TLE, pH 8.0
- 10 mM Tris·Cl, pH 8.0
- 0.1 mM EDTA
- Store at room temperature; stable for years when properly used and tightly closed.
Tris/borate/EDTA buffer (TBE), 10×
- 1 M Tris base
- 1 M boric acid
- 20 mM EDTA, pH 8.0
- Store at room temperature; stable for years when properly used and tightly closed.
Tween wash buffer (TWB)
- 5 mM Tris·Cl, pH 8.0
- 0.5 mM EDTA
- 1 M NaCl
- 0.05% Tween (Fisher, cat. no. 9005-64-5)
- Store at room temperature; stable for years when properly used and tightly closed.
COMMENTARY
Background Information
Proximity-ligation-based chromosome conformation capture techniques, whether 3C, circular chromatin conformation capture (4C), chromosome conformation capture carbon copy (5C), Hi-C, or other adaptations, detect interaction frequencies between any two genomic loci (Dekker et al., 2002; Dostie et al., 2006; Lieberman-Aiden et al., 2009; Simonis et al., 2006). Hi-C was the first of these techniques to map chromosome conformation genome-wide (Lieberman-Aiden et al., 2009). This was achieved by replacing the traditional PCR readout with genome-wide paired-end sequencing. The use of 4-bp-cutting restriction enzymes as in current Hi-C protocols (Belaghzal, Dekker, & Gibcus, 2017) or micrococcal nuclease as in Micro-C (Hsieh et al., 2015; Krietenstein et al., 2020) increased the resolution of chromatin interaction maps as compared to earlier Hi-C assays, which employed 6-bp-cutting restriction enzymes (Lieberman-Aiden et al., 2009). For generating very-high-resolution human genome interaction maps (i.e., with 1-kb bins), Micro-C is a great alternative to Hi-C (Hsieh et al., 2015; Krietenstein et al., 2020; Akgol Oksuz et al., 2020). However, in a recent comparison, we have shown that although Micro-C is superior for detecting (short-range) loops, conventional Hi-C is better suited for quantitative compartment detection. Moreover, Micro-C requires a tight titration of MNase to ensure that there is sufficient digestion in both euchromatin and heterochromatin (A and B compartments; Krietenstein et al., 2020), and it cannot be used for organisms without nucleosomes. Here, we combine DdeI and DpnII to generate smaller fragments than are produced by either enzyme alone (Fig. 6). An important observation to note is that theoretical in silico digestion generates fragments even smaller than those observed in practice after actual digestion (Fig. 6). We refer to this difference as the enzyme penetration. Enzyme penetration effects are expected for most, if not all, enzymes or enzyme combinations, yet the larger the number of restriction sites (i.e., the smaller the expected fragments), the greater the chance of encountering undigested sites. We note that this is not the result of insufficient enzyme activity, as the addition of a second batch of enzymes, at step 14 of Basic Protocol 2, does not improve digestion (data not shown).
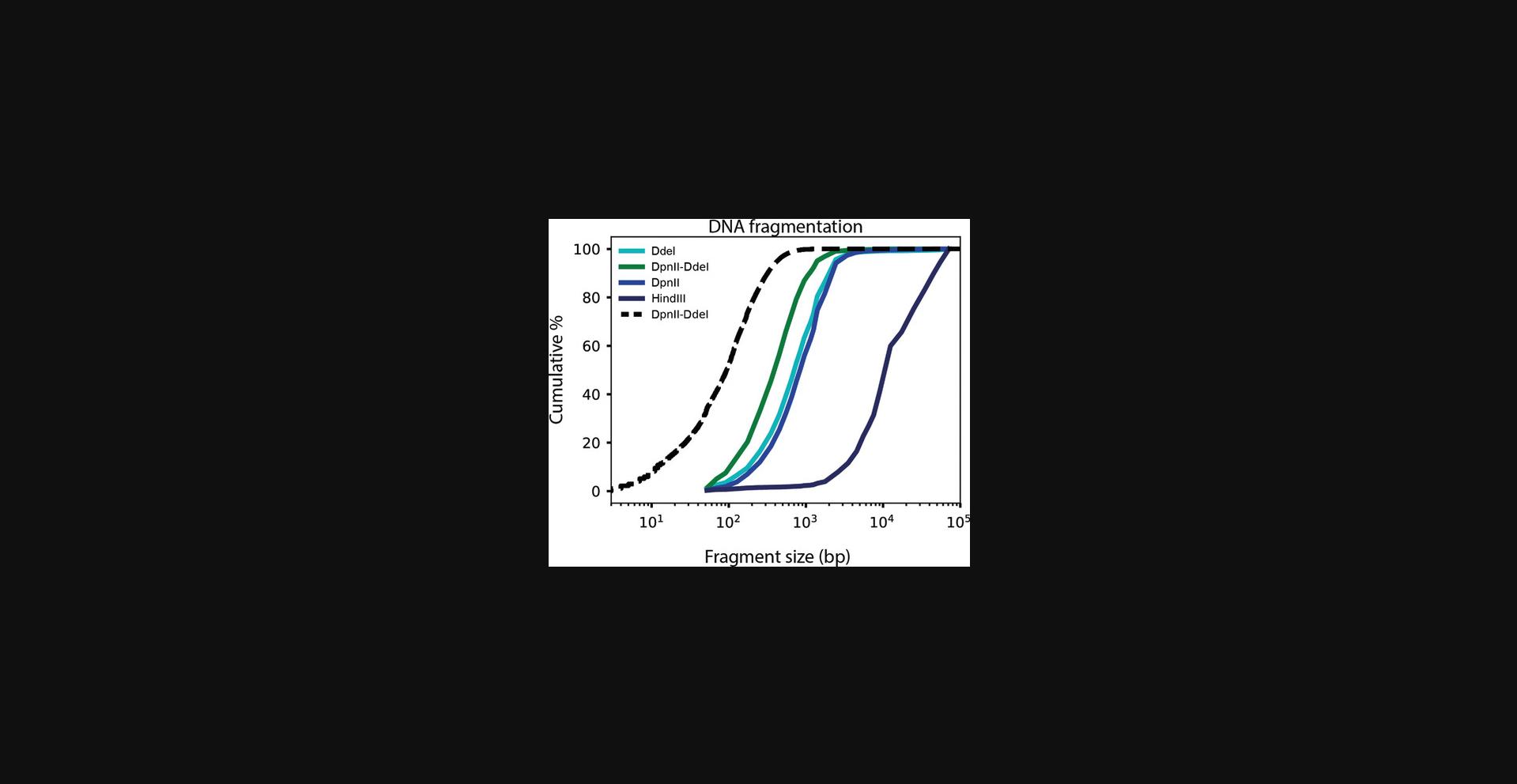
Combinations of chemical fixatives have previously been used in assays such as chromatin immunoprecipitation (ChIP; Zeng, Vakoc, Chen, Blobel, & Berger, 2006), chromatin interaction analysis by paired-end tag sequencing (ChIA-PET; Fullwood et al., 2009), and Micro-C (Hsieh et al., 2015). We have recently shown that Hi-C protocols benefit from adding extra cross-linkers (Akgol Oksuz et al., 2020). The combined use of an additional cross-linker and increased restriction digestion (as described here) provides a Hi-C protocol that simultaneously maximizes the quantitative detection of both large (megabase-scale) features such as compartments and smaller (kilobase-scale) features such as TADs and loops.
Critical Parameters
Preparation and fixation of cells
We recommend harvesting ∼5 × 106 cells to perform this assay, but the protocol has successfully been used with 1-10 × 106 cells as input. Cells are typically counted before fixation, and although we do expect a certain degree of cell loss after fixation, this has been factored into the protocol. Another important note is that the protocol may not be as robust to specific cell types and cell purity. For instance, homogenized hepatic tissue can be rich in lipids, and additional washes may be required before fixation when using samples from such tissue. In general, it is important to collect cells or tissue in a way that minimizes interference with fixation. Improper fixation can increase background signal by resulting in the capture of interactions with randomly floating chromatin. A good way to visualize this background is to quantify interactions between mitochondrial and chromosomal DNA. In contrast to trans-chromosomal interactions, these interactions cannot be specific and are the result of random ligation events between loci that were not cross-linked (Akgol Oksuz et al., 2020). Improper mixing of your fixative solution can create a concentration gradient that can result in cell-to-cell differences in fixation (Golloshi, Sanders, & McCord, 2018). Serum components such as BSA in cell growth media can also negatively affect formaldehyde cross-linking, and these should be thoroughly washed out before cross-linking (Belton et al., 2012). Included in this protocol is the implementation of DSG which, in addition to the traditionally used formaldehyde, greatly improves Hi-C data quality by reducing random ligation events and thereby increasing signal-to-noise ratios (Akgol Oksuz et al., 2020).
Capturing chromosome conformation
For successful conformation capture to occur, chromatin needs to be (1) held in place by fixation, (2) cut into desired fragments, (3) marked with a selectable moiety (i.e., biotin), and (4) ligated to DNA in close 3D proximity. Between the digestion, biotin fill-in, and ligation steps, samples should be kept on ice to avoid reversal of cross-links, which would disrupt chromatin interactions. Keeping samples chilled also minimizes heat-inactivation of enzymes. In preparation for digestion, samples are briefly incubated at 65°C in the presence of SDS to open up the chromatin and increase restriction enzyme accessibility. Chromatin is then digested into fragments of a desired length. In silico digestion can be used to select the enzyme(s) of choice, but enzyme penetration will ultimately determine the quality and resolution (see Background Information; Fig. 6). DpnII-DdeI can be replaced with other enzymes, such as HindIII, which would be preferred if compartments are the feature of interest (Akgol Oksuz et al., 2020); however, altering restriction enzymes may require altering the restriction enzyme buffer used (for instance, we use NEBuffer 2.1 for HindIII). Another important note is that restriction enzymes that create a 5′ overhang must be used to enable Klenow-mediated fill-in. The use of potassium-acetate-containing buffers such as NEBuffer 4/CutSmart buffer (NEB) should be avoided, as they form SDS-potassium precipitates upon the addition of the lysed cell mixture (Umbarger, 2012). Also, depending on the overhang generated by the restriction enzyme, different biotinylated dNTPs may be required (we use biotin-14-dCTP for HindIII). We note that ligation efficiency may be reduced when the biotin is located at the 3′ end of the overhang.
The fragment ends resulting from DpnII and DdeI digestion are blunted in the presence of biotinylated dATP, allowing ligated junctions to be selectively pulled down with streptavidin-coated beads. The fill-in of digested fragment ends needs to be extended from 15 min at 25°C—which is recommended by the manufacturer (New England Biolabs)—to 4 hr at 23°C, due to the bulky biotin moiety. Ligation of the blunted, filled-in ends marks the last critical step of conformation capture. Besides ligation products that arise due to spatial proximity, other ligation events can also form. These include self-circles, dangling ends, and random ligation events between fragments that were floating (i.e., not cross-linked). Self-circles and dangling ends are reduced by performing proximity ligation in situ, which was originally adapted in previous versions of the Hi-C protocol (Belaghzal et al., 2017; Rao et al., 2014). The addition of DSG to 1% formaldehyde, as detailed here, has been shown to reduce trans interactions between floating DNA fragments (Akgol Oksuz et al., 2020).
Preparing Hi-C libraries for sequencing
To selectively recover proximally ligated junctions, biotin incorporated into unligated fragment ends must be removed (Belaghzal et al., 2017). After biotin removal from unligated ends, the Hi-C library can be reduced to a length that is optimal for Illumina paired-end sequencing. In our experience, sonication using Covaris machines yields a reproducibly narrow-size distribution of fragments that is then further narrowed down by size selection using AMPure XP beads. After size selection, DNA fragment ends are repaired and blunted, and biotinylated fragments are bound to streptavidin-coated magnetic beads, which facilitates downstream processing of the library. dATP is then incorporated into the 3′-end of fragment ends, enabling ligation of sequencing adapters. As PCR is used to amplify the final library, a proofreading enzyme should be employed to minimize sequence alterations.
Troubleshooting
Assessing digestion, fill-in, and ligation after DNA isolation
The first quality control step occurs after DNA isolation, when the fragment size distributions for chromatin integrity controls (CI), digestion controls (DC), and proximity ligation samples are assessed (Fig. 2). The CI control should yield a thick band that runs higher than 10 kb for large (mammalian) genomes. Compromised chromatin integrity can manifest as a smear of DNA rather than a narrow band above 10 kb observed with mammalian genomic preparations. Smearing can arise from endogenous nuclease activity before fixation or from slow or delayed freezing of cells after fixation, or can occur if samples have been stored in the freezer for too long before processing. To avoid chromatin integrity issues, fixed cells should be snap-frozen and stored at −80°C, preferably for no longer than 6 months. Cells should also be lysed in sufficient concentrations of protease inhibitor cocktail.
The DC should yield a range of sizes on the gel that reflects the expected fragmentation distribution. Again, note that this size distribution will typically be larger than the expected range determined by in silico digestion because of incomplete enzyme penetration (see Background Information). If restriction digestion does not produce a smear of expected fragment sizes, enzymatic activity was likely inhibited. When repeating the experiment, make sure that buffers and enzymes are fresh and are at the correct concentration and pH. For example, the use of 10% instead of 1% SDS stock solution to open up DNA will completely abolish restriction enzyme activity. Underestimating the input cell number for the assay can also reduce chromatin digestion efficiency.
Finally, because the proximity ligation sample is the product of ligation events that occur between digested fragments, the “Hi-C sample” on the gel should have a higher average molecular weight distribution than the DC. Loading several dilutions of the Hi-C sample together with a known amount of DNA ladder provides a means to quantify the DNA yield. Hi-C experiments typically yield >20 µg of DNA at this point, and <2 µg of DNA may be insufficient to carry over for Illumina library preparation. The most common reason that Hi-C experiments fail is because there is no observable upward shift in molecular weight between the digestion and proximity ligation, and regrettably, this is also the most difficult scenario to troubleshoot. Any number of deficiencies and/or inefficiencies between cell preparation and proximity ligation can produce a failed ligation. One option would be to design a PCR-based strategy that samples one specific chromatin interaction to assess whether fill-in and ligation occurred with both enzymes (Fig. 7).
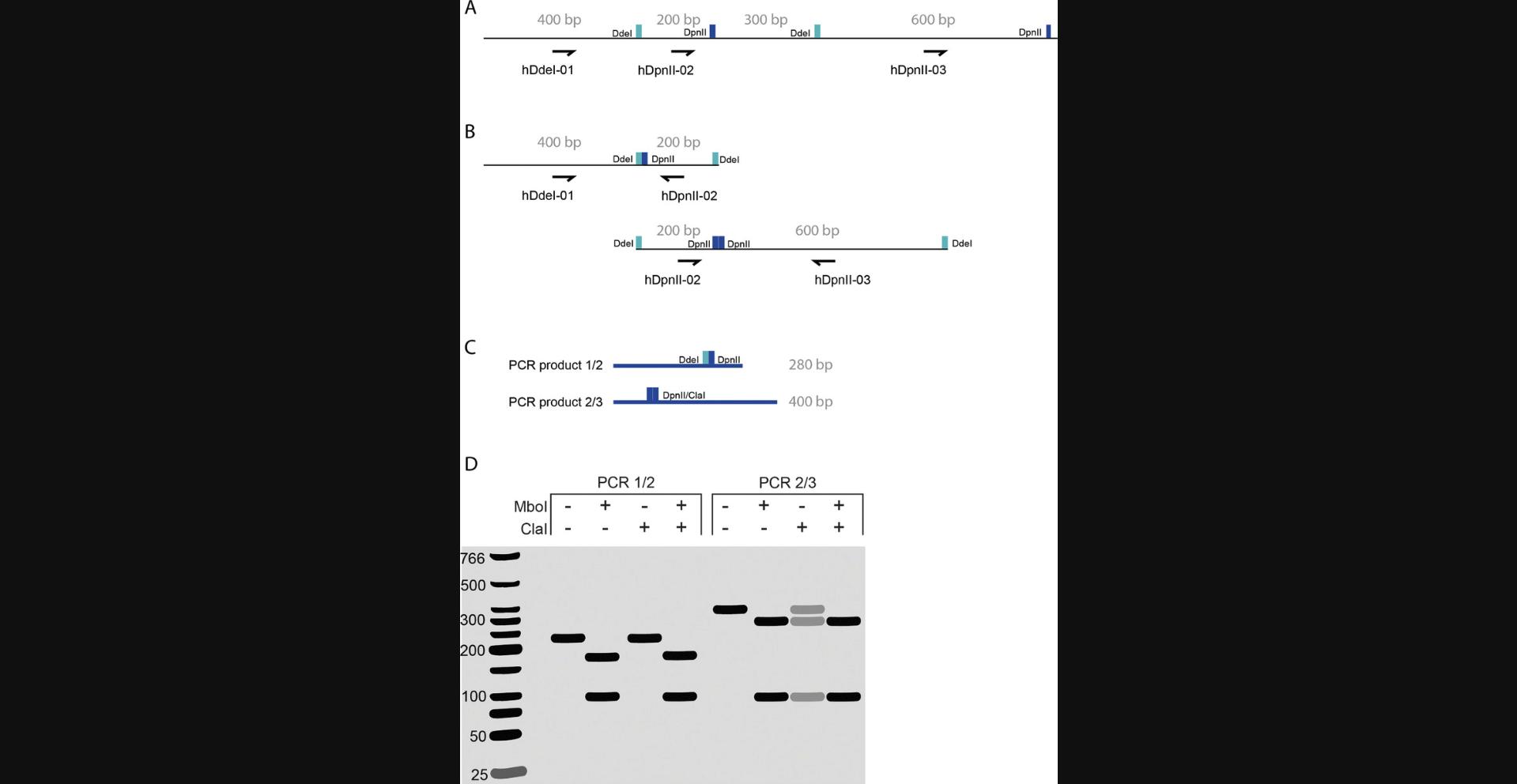
Another logical step would be to repeat the experiment with fresh buffers and enzymes on the correct number of fresh cells. If all of these troubleshooting approaches fail, we would recommend performing the conformation capture reactions on naked DNA. To this end, genomic DNA is isolated either immediately before or immediately after chromatin digestion. Purified DNA can then be carried forward to digestion, fill-in, and ligation (for DNA isolated before chromatin digestion) or fill-in and ligation (for DNA isolated after chromatin digestion). The digestion, fill-in, and ligation products can then be subjected to gel electrophoresis. Because digestion, fill-in, and ligation are more efficient on naked DNA, this test could identify problems arising when performing the reactions in situ. Note that these troubleshooting steps are not performed on fixed cells and, therefore, will not produce an actual Hi-C library.
From DNA to size selection
After chromosome conformation is captured through proximity ligation, the sample is much less vulnerable but can still be lost in various ways. When using size-selection columns or AMPure beads, it is good practice to not discard any beads, supernatants, or column flowthrough. If your sample is not apparent on a gel, it must be in one of these fractions and can most often be recovered. If, after size selection, the DNA from the Lower Fraction cannot be recovered or is of insufficient quantity, DNA from the Upper Fraction can be re-sonicated to produce more of the Lower Fraction. Gel quantification will determine whether the sonication was efficient and if enough DNA was recovered.
Biotin pulldown to PCR
After size selection, the next quality assessment point is the titration PCR. If the titration PCR fails, the problem can typically be traced back and troubleshot relatively easily. If fill-in and ligation are successful, and DNA quantification of the library was accurate (see Fig. 2), then you know you have generated a proximally ligated library. Unless you mistakenly used dATP instead of biotin-dATP for biotin fill-in, DNA must be bound to your streptavidin-coated beads. Therefore, the first step in troubleshooting issues with titration PCR would be to redo the end-repair, A-tailing, and adapter ligation steps. If this fails, we advise starting from the Upper Fraction that was isolated from AMPure beads, as explained in the previous subsection, “From DNA to size selection.”
Production PCR and cleanup
Once the optimal amount of PCR cycles is determined (see Fig. 4), a production PCR is performed to generate final sequencing libraries. Production PCR libraries should yield a smear similar to that generated by the titration PCR that is, on average, the size of the Lower AMPure fraction plus adaptors. The last step is to remove primer dimers from the final libraries. If primer dimers are highly abundant, two consecutive rounds of AMPure purification may be required.
Understanding Results
A typical sequencing result on Hi-C libraries produced following the protocols outlined here, for the human genome, will have ∼50% of reads uniquely mapped, of which >70% are expected to be interchromosomal (cis), but this is very much cell type dependent (see Table 3). As PCR duplicates (exact sequence matches for a given ligated fragment) are discarded during analysis and contribute to increased sequencing cost, they should be limited to <15%. Note that not all duplicates detected by sequencing are PCR duplicates; some duplication can occur on the Illumina flow cells, which are referred to as optical duplicates. When performed on human cells, this protocol should provide enough DNA for at least one lane of sequencing, and 6-7 PCR cycles is typically sufficient to amplify libraries (see titration PCR). However, in the past we have successfully mapped libraries generated with 14 PCR cycles without excessive amounts of PCR duplicates.
| Reads | Number | % |
|---|---|---|
| Total reads | 6,588,364,593 | 100.00 |
| Unmapped | 477,936,271 | 7.25 |
| Single side mapped | 1,870,513,027 | 28.39 |
| Both sides mapped | 4,239,915,295 | 64.35 |
| Duplications (PCR) | 960,922,809 | 14.59 |
| Total with no duplications (valid pairs) | 3,278,992,486 | 49.77 |
| Cis | 2,930,159,461 | 89.36 |
| Trans | 348,833,025 | 10.64 |
- a Note that this is the amount of reads derived from three flow cells on an Illumina HiSeq4000 sequencer.
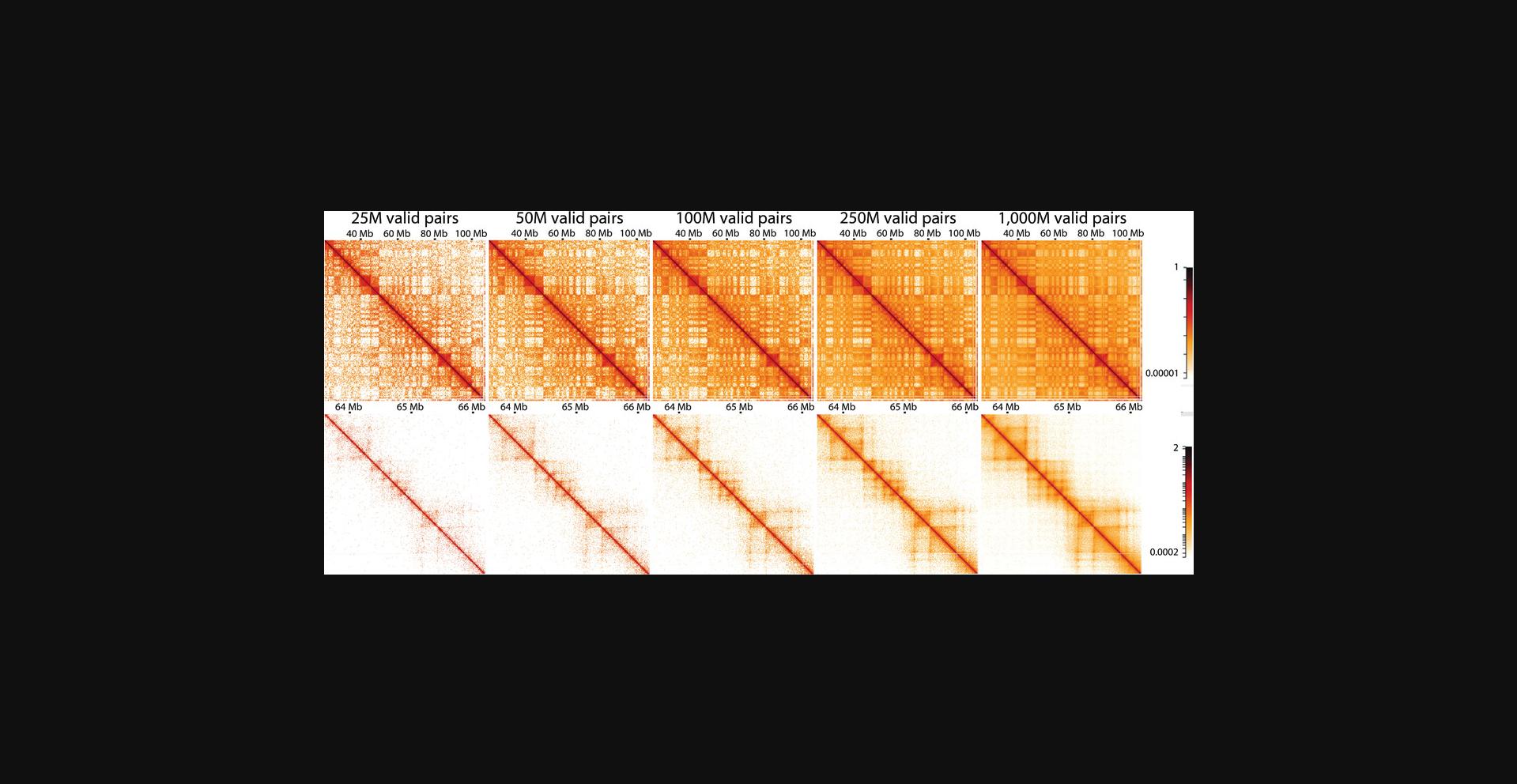
Because of the polymeric nature of DNA, there is a distance-dependent decay of interaction frequency. This relation is best depicted by plotting the interaction frequency over genomic distance (Lajoie et al., 2015), but it can also be observed directly from Hi-C matrices, as interactions become more infrequent further away from the central diagonal (Fig. 8). Many genomic folding principles, such as compartments, TADs, and loops, can be identified as deviations from this general distance-dependent decay. As discussed in the Strategic Planning section, sequencing depth is an important consideration when planning an experiment, as libraries should be sequenced at a depth at which structures of interest can be resolved. For example, large structures such as compartments in human cells can be resolved when plotting only 20-25 million valid pairs, whereas smaller structures such as TADs and loops require much higher resolution and, thus, many more reads (for loops, >1 billion reads are recommended; Fig. 8).
Time Considerations
The generation of Hi-C libraries ready for sequencing can comfortably be completed in 6 days regardless of how many samples are being generated. However, the protocol can be condensed to as little as 4 days, especially if few samples are being processed.
Cell fixation after harvest can be performed in a few hours. Given the long incubations for digestion, fill-in, and ligation, generating raw Hi-C libraries (Basic Protocol 2) from fixed cells is restricted to 3 days. The end of Basic Protocol 2 marks the point at which samples can be stored long term at −20°C or short term at 4°C at any point within the protocol, with the exception that adapter ligation should immediately follow A-tailing. Biotin removal can be performed immediately after DNA purification, by running samples in a thermocycler overnight and recovering them the next day, if the program is terminated with a refrigerated hold step.
Some ambitious users have combined DNA purification (Basic Protocol 2) and biotin pulldown (Basic Protocol 3) in a single day, which would permit the remainder of Basic Protocol 3 to be completed the next day. Once the biotinylated fragments are bound to streptavidin-coated beads, final Hi-C libraries can be generated within 1-3 days, but the time required is largely dependent on the number of samples being processed simultaneously. To this end, it is worth considering how many sample spaces are available on the MPS or rotators, which may limit the number of samples that can be processed in parallel. Using the materials listed in this article, experienced users should be able to comfortably process 16 samples simultaneously. One way to process more samples would be to start from fixed cells that are frozen and process two batches of samples on consecutive days. Here, while the second batch is digesting, the first batch can easily be filled in and ligated. The next day, the DNA purification of the first batch can feasibly be combined with fill-in and ligation of the second batch. Afterward, both batches can be processed simultaneously.
Acknowledgments
The authors thank Hakan Ozadam for analyzing the enzyme penetration data, and Betul Akgol Oksuz, Nils Krietenstein, and Sameer Abraham for their involvement and analysis of the Hi-C 3.0 protocol data. The authors acknowledge support from the US National Institutes of Health (NIH) Common Fund 4D Nucleome Program (U54DK107980) and the Ruth L. Kirschstein Predoctoral Individual National Research Service Award (F31HG011583).
Author Contributions
Denis L. Lafontaine : Methodology, Writing-original draft, Writing-review & editing; Liyan Yang : Methodology, Writing-original draft, Writing-review & editing; Job Dekker : Supervision; Johan H. Gibcus : Supervision, Writing-original draft, Writing-review & editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Akgol Oksuz, B., Yang, L., Abraham, S., Venev, S. V., Krietenstein, N., Parsi, K. M., … Dekker, J. (2020). Systematic evaluation of chromosome conformation capture assays [Preprint]. BioRxiv , 2020.12.26.424448. Retrieved from https://www.biorxiv.org/content/10.1101/2020.12.26.424448v2
- Belaghzal, H., Dekker, J., & Gibcus, J. H. (2017). Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods , 123, 56–65. doi: 10.1016/j.ymeth.2017.04.004.
- Belton, J.-M., & Dekker, J. (2015). Hi-C in budding yeast. Cold Spring Harbor Protocols , 2015, 649–661. doi: 10.1101/pdb.prot085209.
- Belton, J.-M., McCord, R. P., Gibcus, J. H., Naumova, N., Zhan, Y., & Dekker, J. (2012). Hi-C: A comprehensive technique to capture the conformation of genomes. Methods , 58, 268–276. doi: 10.1016/j.ymeth.2012.05.001.
- Dekker, J., Rippe, K., Dekker, M., & Kleckner, N. (2002). Capturing chromosome conformation. Science , 295, 1306–1311. doi: 10.1126/science.1067799.
- Dostie, J., Richmond, T. A., Arnaout, R. A., Selzer, R. R., Lee, W. L., Honan, T. A., … Dekker, J. (2006). Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Research , 16, 1299–1309. doi: 10.1101/gr.5571506.
- Fullwood, M. J., Liu, M. H., Pan, Y. F., Liu, J., Xu, H., Mohamed, Y. B., … Ruan, Y. (2009). An oestrogen-receptor-alpha-bound human chromatin interactome. Nature , 462, 58–64. doi: 10.1038/nature08497.
- Golloshi, R., Sanders, J. T., & McCord, R. P. (2018). Iteratively improving Hi-C experiments one step at a time. Methods , 142, 47–58. doi: 10.1016/j.ymeth.2018.04.033.
- Hoffman, E. A., Frey, B. L., Smith, L. M., & Auble, D. T. (2015). Formaldehyde crosslinking: A tool for the study of chromatin complexes. Journal of Biological Chemistry , 290, 26404–26411. doi: 10.1074/jbc.R115.651679.
- Hsieh, T.-H. S., Weiner, A., Lajoie, B., Dekker, J., Friedman, N., & Rando, O. J. (2015). Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell , 162, 108–119. doi: 10.1016/j.cell.2015.05.048.
- Krietenstein, N., Abraham, S., Venev, S. V., Abdennur, N., Gibcus, J., Hsieh, T.-H. S., … Rando, O. J. (2020). Ultrastructural details of mammalian chromosome architecture. Molecular Cell , 78, 554–565.e7. doi: 10.1016/j.molcel.2020.03.003.
- Lajoie, B. R., Dekker, J., & Kaplan, N. (2015). The hitchhiker's guide to Hi-C analysis: Practical guidelines. Methods , 72, 65–75. doi: 10.1016/j.ymeth.2014.10.031.
- Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., … Dekker, J. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science , 326, 289–293. doi: 10.1126/science.1181369.
- Marinov, G. K., Trevino, A. E., Xiang, T., Kundaje, A., Grossman, A. R., & Greenleaf, W. J. (2021). Transcription-dependent domain-scale three-dimensional genome organization in the dinoflagellate Breviolum minutum. Nature Genetics , 53, 613–617. doi: 10.1038/s41588-021-00848-5.
- Moissiard, G., Cokus, S. J., Cary, J., Feng, S., Billi, A. C., Stroud, H., … Jacobsen, S. E. (2012). MORC family ATPases required for heterochromatin condensation and gene silencing. Science , 336, 1448–1451. doi: 10.1126/science.1221472.
- Moore, D., & Dowhan, D. (2002). Purification and concentration of DNA from aqueous solutions. Current Protocols in Molecular Biology , 59, 2.1A.1–2.1A.10. doi: 10.1002/0471142727.MB0201AS59.
- Nand, A., Zhan, Y., Salazar, O. R., Aranda, M., Voolstra, C. R., & Dekker, J. (2021). Genetic and spatial organization of the unusual chromosomes of the dinoflagellate Symbiodinium microadriaticum. Nature Genetics , 53, 618–629. doi: 10.1038/s41588-021-00841-y.
- Rao, S. S. P., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., … Aiden, E. L. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell , 159, 1665–1680. doi: 10.1016/j.cell.2014.11.021.
- Simonis, M., Klous, P., Splinter, E., Moshkin, Y., Willemsen, R., de Wit, E., … de Laat, W. (2006). Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature Genetics , 38, 1348–1354. doi: 10.1038/ng1896.
- Strang, C., Cushman, S. J., DeRubeis, D., Peterson, D., & Pfaffinger, P. J. (2001). A central role for the T1 domain in voltage-gated potassium channel formation and function. Journal of Biological Chemistry , 276, 28493–28502. doi: 10.1074/jbc.M010540200.
- Umbarger, M. A. (2012). Chromosome conformation capture assays in bacteria. Methods , 58, 212–220. doi: 10.1016/j.ymeth.2012.06.017.
- Wang, X., Le, T. B. K., Lajoie, B. R., Dekker, J., Laub, M. T., & Rudner, D. Z. (2015). Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes & Development, 29, 1661–1675.
- Zeng, P.-Y., Vakoc, C. R., Chen, Z.-C., Blobel, G. A., & Berger, S. L. (2006). In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques , 41, 694, 696, 698. doi: 10.2144/000112297.
Citing Literature
Number of times cited according to CrossRef: 15
- Xiaoyuan Tao, Sujuan Li, Guang Chen, Jian Wang, Shengchun Xu, Approaches for Modes of Action Study of Long Non-Coding RNAs: From Single Verification to Genome-Wide Determination, International Journal of Molecular Sciences, 10.3390/ijms24065562, 24 , 6, (5562), (2023).
- Evelyn Kabirova, Artem Nurislamov, Artem Shadskiy, Alexander Smirnov, Andrey Popov, Pavel Salnikov, Nariman Battulin, Veniamin Fishman, Function and Evolution of the Loop Extrusion Machinery in Animals, International Journal of Molecular Sciences, 10.3390/ijms24055017, 24 , 5, (5017), (2023).
- Thomas John Lopdell, Using QTL to Identify Genes and Pathways Underlying the Regulation and Production of Milk Components in Cattle, Animals, 10.3390/ani13050911, 13 , 5, (911), (2023).
- Jingjing Li, Haiyan Wang, Jianqing Zhu, Qi Yang, Yang Luan, Leming Shi, José Arturo Molina-Mora, Yuanting Zheng, De novo assembly of a chromosome-level reference genome of the ornamental butterfly Sericinus montelus based on nanopore sequencing and Hi-C analysis, Frontiers in Genetics, 10.3389/fgene.2023.1107353, 14 , (2023).
- Ananda L. Roy, Richard S. Conroy, Veronica G. Taylor, Judy Mietz, Ian M. Fingerman, Michael J. Pazin, Phillip Smith, Carolyn M. Hutter, Dinah S. Singer, Elizabeth L. Wilder, Elucidating the structure and function of the nucleus—The NIH Common Fund 4D Nucleome program, Molecular Cell, 10.1016/j.molcel.2022.12.025, 83 , 3, (335-342), (2023).
- Chi-Nga Chow, Chien-Wen Yang, Wen-Chi Chang, Databases and prospects of dynamic gene regulation in eukaryotes: A mini review, Computational and Structural Biotechnology Journal, 10.1016/j.csbj.2023.03.032, 21 , (2147-2159), (2023).
- Nan Zhang, Shruthi Kandalai, Xiaozhuang Zhou, Farzana Hossain, Qingfei Zheng, Applying multi‐omics toward tumor microbiome research, iMeta, 10.1002/imt2.73, 2 , 1, (2023).
- Xiaolin Wei, Duc Tran, Yarui Diao, HiCAR: Analysis of Open Chromatin Associated Long‐range Chromatin Interaction Using Low‐Input Materials, Current Protocols, 10.1002/cpz1.899, 3 , 10, (2023).
- Oleg V. Bylino, Airat N. Ibragimov, Filomena Anna Digilio, Ennio Giordano, Yulii V. Shidlovskii, Application of the 3C Method to Study the Developmental Genes in Drosophila Larvae, Frontiers in Genetics, 10.3389/fgene.2022.734208, 13 , (2022).
- Xuelong Wang, Jizhou Yan, Zhao Ye, Zhiqiang Zhang, Sheng Wang, Shuang Hao, Baiyong Shen, Gang Wei, Reorganization of 3D chromatin architecture in doxorubicin-resistant breast cancer cells, Frontiers in Cell and Developmental Biology, 10.3389/fcell.2022.974750, 10 , (2022).
- Junhong Wang, Chunwei Shi, Mingyang Cheng, Yiyuan Lu, Xiaoyu Zhang, Fengdi Li, Yu Sun, Xiaoxu Li, Xinyang Li, Yan Zeng, Chunfeng Wang, Xin Cao, Effects of the Zbtb1 Gene on Chromatin Spatial Structure and Lymphatic Development: Combined Analysis of Hi-C, ATAC-Seq and RNA-Seq, Frontiers in Cell and Developmental Biology, 10.3389/fcell.2022.874525, 10 , (2022).
- Satish Sati, Parker Jones, Hali S. Kim, Linda A. Zhou, Emmanuel Rapp-Reyes, Thomas H. Leung, HiCuT: An efficient and low input method to identify protein-directed chromatin interactions, PLOS Genetics, 10.1371/journal.pgen.1010121, 18 , 3, (e1010121), (2022).
- Sara Rodriguez, Ashley Ward, Andrew T Reckard, Yulia Shtanko, Clayton Hull-Crew, Andrew D Klocko, The genome organization of Neurospora crassa at high resolution uncovers principles of fungal chromosome topology , G3 Genes|Genomes|Genetics, 10.1093/g3journal/jkac053, 12 , 5, (2022).
- Kyoung-Jae Choi, My Diem Quan, Chuangye Qi, Joo-Hyung Lee, Phoebe S. Tsoi, Mahla Zahabiyon, Aleksandar Bajic, Liya Hu, B. V. Venkataram Prasad, Shih-Chu Jeff Liao, Wenbo Li, Allan Chris M. Ferreon, Josephine C. Ferreon, NANOG prion-like assembly mediates DNA bridging to facilitate chromatin reorganization and activation of pluripotency, Nature Cell Biology, 10.1038/s41556-022-00896-x, 24 , 5, (737-747), (2022).
- Leina Lu, Fulai Jin, Easy Hi-C: A Low-Input Method for Capturing Genome Organization, DNA-Protein Interactions, 10.1007/978-1-0716-2847-8_9, (113-125), (2022).

