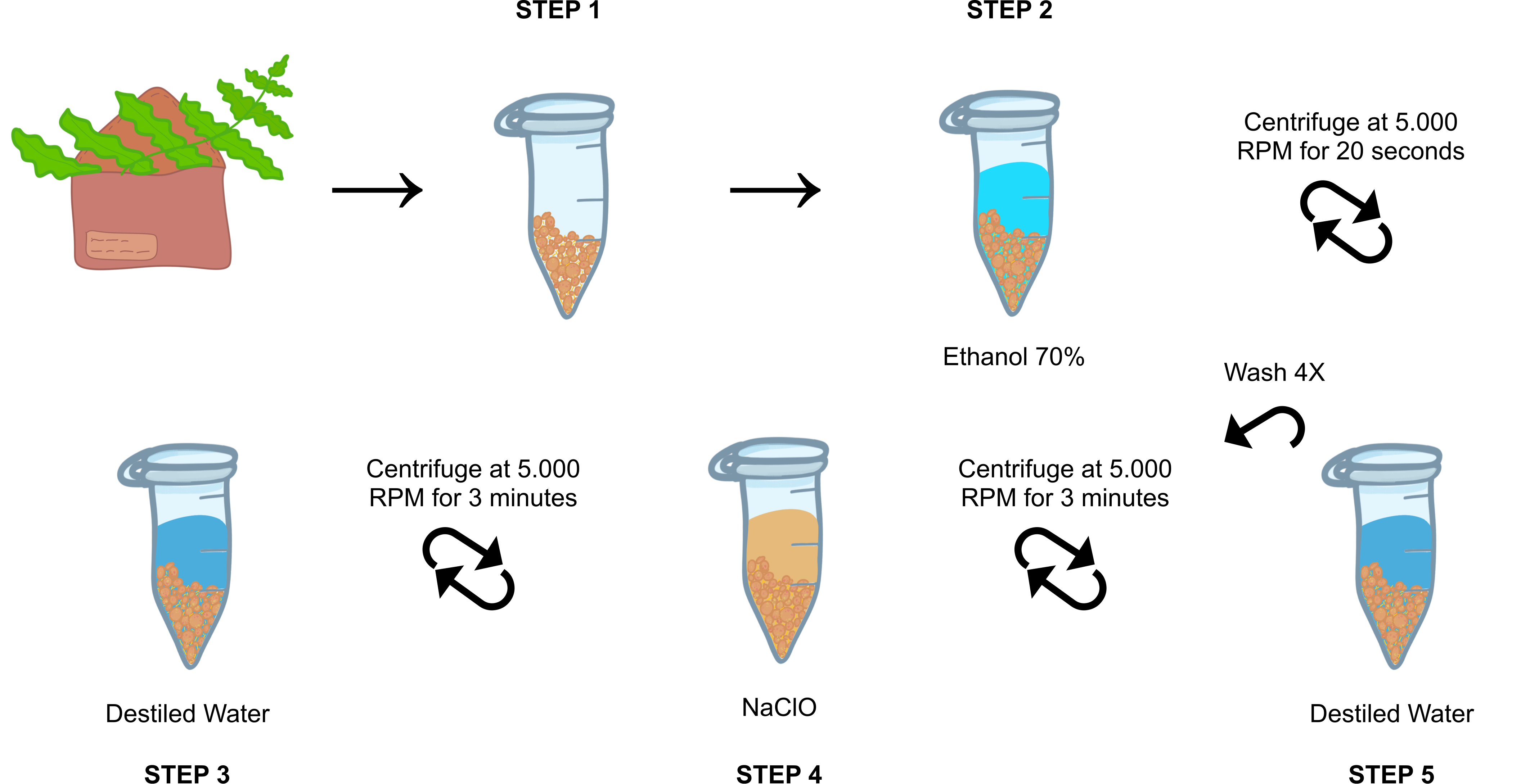HMW DNA extraction protocol for ferns
Geferson Fernando Metz, Rafael Plá Matielo Lemos, Cristiane Barbosa D'Oliveira Matielo, Tiego Ferreira, Filipe Victoria
Abstract
Among the difficulties encountered in a laboratory, simple situations such as efficiently disinfecting fern spores and extracting large amounts of DNA with low tissue input in the protocol are one of the major impediments in carrying out sequencing of these plants in the axenic state. The objective of this work is to present a replicable, scalable and easy-to-execute protocol for work with ferns following the current generation of long-read sequencing. Our method is based on providing a disinfection protocol for spores and sporangia that guarantees growth free of contamination and, after this growth, DNA extraction using a low amount of material in order to obtain a good yield. The results are promising since, in up to 21 days, we obtained germinated plants. After their growth (in average 180 days) we were able to extract DNA in quantity and quality and perform the sequencing, emphasizing that our best N50 is 24 Kb.
Before start
Prepare the SDS Lysis Buffer on the day of the experiment;
| A | B | C |
|---|---|---|
| Reagent | Reagent Stock Concentration | FINAL |
| PVP 40 | 100% | 1% |
| Sodium Metabisulfite | 1% | 1% |
| NaCl | 5 M | 0,5 M |
| TRIS HCL pH 8 | 1 M | 100 mM |
| EDTA pH 8 | 0.5 M | 50 mM |
| DDH2O | - | ~ |
| Sodium dodecyl sulfate (SDS) | 20% | 1,5% |
| β-MERCAPTOETANOL | - | 2% (v/v) |
-
When preparing the SDS Lysis Buffer, add SDS at last this will avoiding bubble formation.
-
Preheat the water bath; Keep the SDS Lysis Buffer at 65°C until the tissue powder is added.
-
Washing solution (EtOH 70%), fresh 35 ml Ethanol 100% + 15 ml H2O
-
Potassium Acetate 5M Dissolve 4.9 gr KAc in 10 ml ddH2O (4.9 gr KAc + ≈ 7.5 ml H2O).
Adjust the pH with glacial acetic acid.
- Prepare TE buffer (10 mM Tris pH 8 and 1 mM EDTA pH 8)
Steps
Plant material sterilization
Sample and store the leaf tissue (fronds) in paper envelopes for three days to induce dehiscence.
Recover and store the spores together with sporangia in 1.5 mL microtubes until one-third of the tube was filled and then stored at -20°C until disinfection.
Add 1mL 0h 0m 20s and briefly centrifuge them.
Discard the supernatant and wash by inversion with 1mL of autoclaved purified water, briefly centrifuge and discard the supernatant.
Add 1mL of an 10% (v/v) , homogenize by inversion for 0h 20m 0s , centrifuge at 5.000rpm,0h 0m 0s 0h 3m 0s.
Discard the supernatant and wash by inversion with 1mL of autoclaved purified water, centrifuge at 55.000rpm,0h 0m 0s 0h 3m 0s. Repeat this step 4 times.
Add 1mL autoclaved purified water, homogenize and pipete into a Petri Dish with BCD medium (MgS04.7H2O - 0.1 mM, KH2PO4 - 1.84 mM, KNO3 - 1M, FeSO4.7H2O - 4.5mM).
Extraction of high-molecular-weight DNA
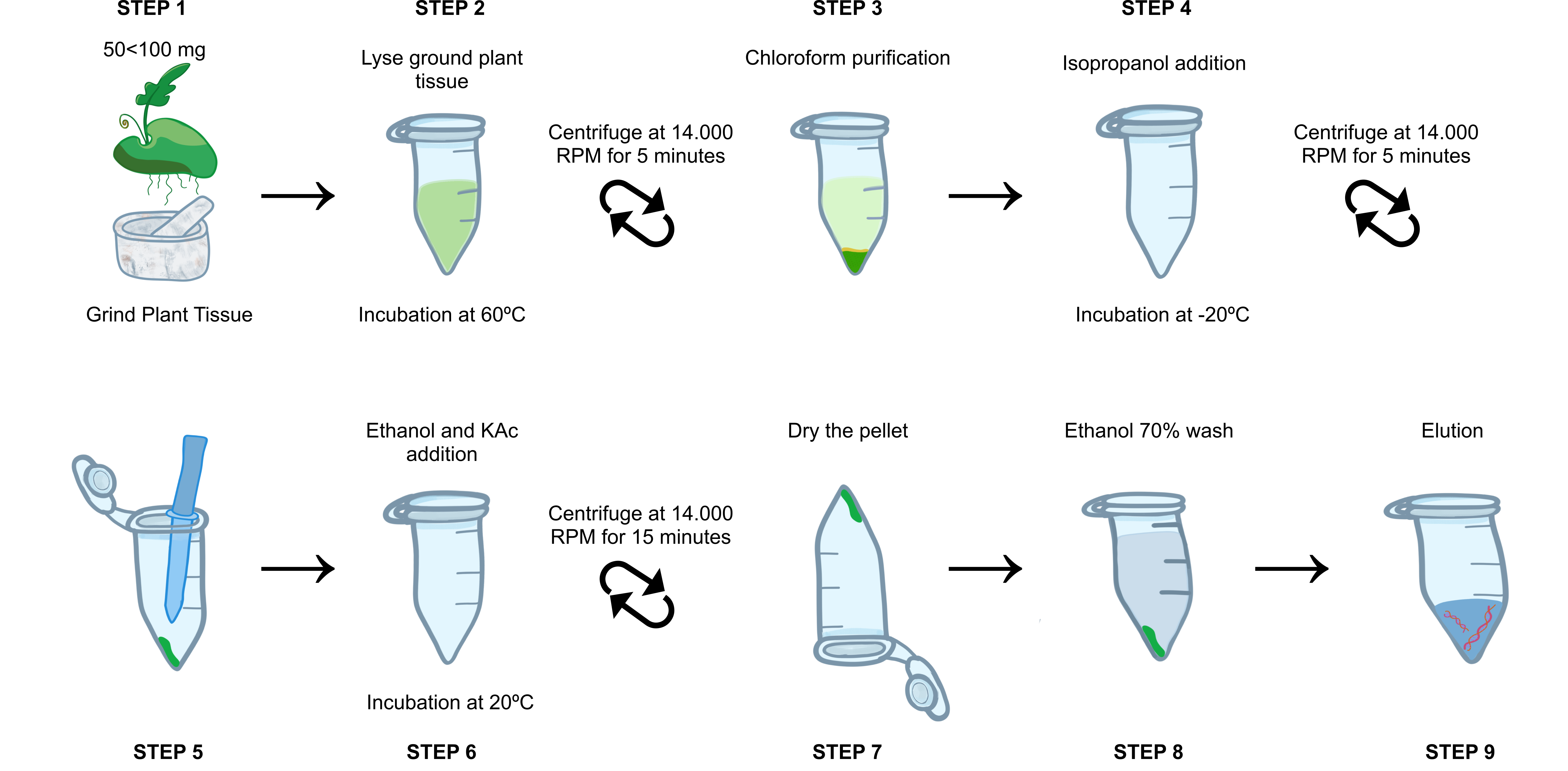
Weigh between 50mgand 100mg of leaf tissue. Grind must be done with a crucible and pestle (previously exposed to
Pre-heat the Lysis Buffer and aliquot 600µLfor each sample individually in 1.5 mL microtube. With the aid of a spoon or spatula, transfer the macerate to the microtube.
Homogenize by inversion 10 times and incubate at 60°C for 0h 10m 0s.
Add 4µLof RNase A (100 mg/mL); add 4µL of proteinase K (> 40 U/mg);
Homogenize by inversion 10 times and incubate at 60°C for 0h 20m 0s gently homogenizing by inversion every 0h 5m 0s.
Add 600µLof 0h 3m 0s until an off-white emulsion forms.
Centrifuge at 14.000rpm,0h 0m 0sfor 0h 5m 0sat room temperature.
Carefully aspirate the upper phase of the tube and transfer to a new 1.5 mL microtube.
Add 400µLof -20°C and incubate it for at least 1h 0m 0s at -20°C. (Can be stored overnight)
Centrifuge at 14.000rpm,0h 0m 0sfor 0h 5m 0s at room temperature and discard the supernatant.
Add 50µLof 5µLof
Add 120µLof iced -20°C) and mix well by flicking the tube.
Store tubes at -20°C for at least 0h 20m 0s.
Centrifuge at 14.000rpm,0h 0m 0s for 0h 15m 0s at room temperature and carefully discard the supernatant.
Add 1mL of freshly prepared
50µL Dry the pellet for approximately 0h 10m 0sat Room Temperature on the bench with the tube upside down.
Elute in 50µL volume in
Quality Assessment
Quantify the DNA on a Qubit® fluorometer (dsDNA high sensitivity assay). DNA yield can be 500 - 1500 ng.
Check DNA integrity though electrophoresis in a 1'% Agarose Gel.
DNA Size Selection
Remove short DNA fragments with Circulomics® Short-Read Eliminator Kit.