HMW DNA extraction for Long Read Sequencing using CTAB
Benoît Vacherie, Karine Labadie, Cyril Falentin
Abstract
High Molecular Weight DNA extraction protocol for Long Read sequencing.
Extraction is performed from flash frozen leaves stored at -80°C.
The protocol is adapted for an extraction of 1g of leaves.
This protocol is based on the protocol provided by Oxford Nanopore Technologies, Oxford, UK (ONT), "High molecular weight gDNA extraction from plant leaves" provided by the ONT community in March 2019, with slighly modifications.
This protocol involves a conventional CTAB extraction followed by purification using commercial Qiagen Genomic tips (QIAGEN, MD, USA). DNA fragment size selection is performed using Short Read Eliminator (Circulomics, MD, USA).
This protocol is particularly adapted for plant leaves, but also works with many other organisms (microalgae, insects...).
Steps
preparation of reagents
Extraction Buffer : Prepare 20 mL of extraction buffer (per 1g of leaves) in a 50 mL tube
| A | B |
|---|---|
| Tris-HCl ph8 | 100 mM |
| EDTA | 20 mM |
| CTAB | 2% |
| NaCl | 1.4. M |
| PEG 8000 | 1 % |
| H2O | QSP 20 ml |
DNA Extraction
Add 50 µl of 2-Mercaptoethanol to 20 ml of extraction buffer and preheat to 65 °C (approx. 15 min).
20mL
50µL
65°C
Put a mortar in ice.
Cool the mortar and pestle with liquid nitrogen until the bubbling stops.
Transfer the powder to the pre-warmed extraction buffer
Add 40 µl of RNase A (100mg/ml) and vortex the tube 5 sec.
40µL
0h 0m 5s
Incubate 1h at 65°C with intermittent agitation (300 rpm every 10 min)
1h 0m 0s
Let the tube cool for 5 min at RT
0h 5m 0s
Add 1 volume (20ml) of chloroform and vortex 2x5sec.
20mL
Gently transfer the aqueous phase to a new 50 ml tube.
Add 0.7 volume of isopropanol and mix gently by inversion 10 times
Put the tube at -80°C for 15 minutes.
0h 15m 0s
If a DNA medusa appears after this step, recover the medusa using a Pasteur pipette (without breaking the tip of the pipette).
Wash the medusa in 3 successive baths of 70% ethanol then resuspend the medusa in 100µl of 1X TE.
Incubate for 1 hour at 55°C then store the tube at 4°C before quality control.
Continue the protocol with the rest of the tube (without the medusa).
If no DNA medusa is visible continue the protocol.
Centrifuge the tube with low acceleration and deceleration
5500x g,4°C
Carrefully remove the supernatant without resuspending the pellet.
Remove the remaining liquid by turning the tube upside down on a paper towel (make sure the pellet does not come off).
Gently resuspend with the pipette the DNA pellet with 9.5 ml of G2 buffer (QIAGEN Genomic-tip 100/G).
Incubate 15 min at 50°C . The sample can be stored overnight at 4°C at this stage.
0h 15m 0s
Génomic tip purification
Equilibrate a QIAGEN Genomic-tip 100/G column with 4 ml of QBT buffer .
Preheat 5 ml of QF buffer to 55°C.
Allow all the QBT buffer to drain by gravity flow into a 50 ml tube.
Add the sample in G2 buffer (9.5ml) on the column and let it to enter the resin by gravity flow.
Wash the column twice with 7.5 ml of QC buffer.
Put the column on a 15 ml tube.
Elute the DNA with 5 ml of QF buffer preheated to 55°C.
DNA précipitation
Add 0.7 volume of isopropanol to the eluate.
Mix gently by inverting 20 times.
Incubate 15 minutes at RT.
0h 15m 0s
Centrifuge with low acceleration and deceleration
5500x g,4°C
Carefully discard the supernatant, gently resuspend the pellet with 1 ml of cold 70% ethanol.
Transfer the resuspended DNA to a 1.5 ml DNA LoBind tube.
Centrifuge with low acceleration and deceleration
5000x g,4°C
Carefully discard the supernatant.
Air dry the pellet at RT for about 10 minutes.
0h 10m 0s
Resuspend the pellet with 50-100 µl of 1X TE buffer.
Allow the DNA to resuspend for 2 hours at 55°C or ON at RT.
2h 0m 0s or
Store the DNA at 4°C.
Sample quality control
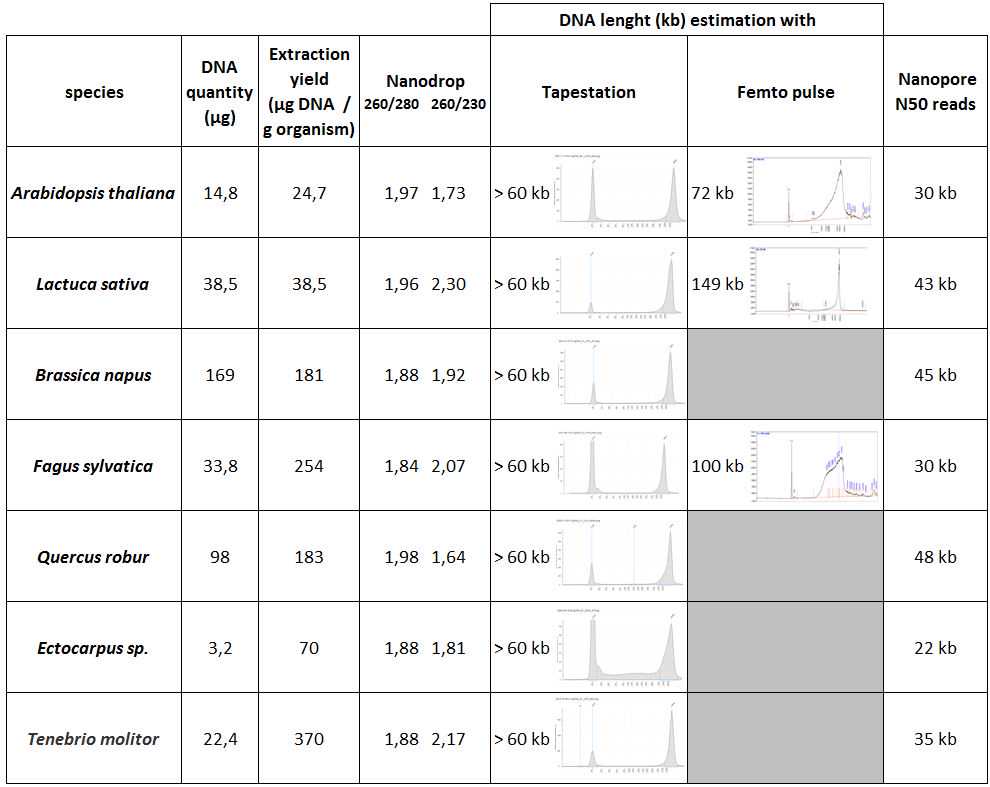
Quantify your sample with a Qubit DNA HS assay.
Check the purity of the sample with a Nanodrop (measurements of 260/280 and 260/230 absorbance ratios).
Estimate the molecular weight of the sample with a Tapestation and/or a Femto pulse and/or a Pippin Pulse.
Depending on the DNA concentrations and DNA length profiles, deplete short DNA molecules using SRE size selection kits (SRE XS, SRE or SRE XL kits).