Golgi immunopurification (Golgi-IP) for subcellular lipid profiling
Wentao Dong, Eshaan S Rawat, Monther Abu-Remaileh
Abstract
The Golgi is a membrane-bound organelle that is central to protein and lipid processing, sorting and secretion in the cell. Despite its critical cellular function, there has been challenges to quantitatively assess Golgi lipid profiles. To overcome this hurdle, we developed a rapid harvesting and purification method using immunoprecipitation (Golgi-IP). This protocol provides details for preparing Golgi-IP lipidomics samples.
Steps
Preparation of homogenizers and sample tubes
Wash the glass vessel homogenizer with MilliQ Water, 10 times each. Wash the tissue grinder homogenizer thoroughly with DI Water and MilliQ Water, especially the gap between the white parts, don’t touch the part that goes into the glass vessel. Then dry upside-down using paper towels. Carefully place the glass vessels against something to prevent falling down. Minimize any contact between the grinder and anything else.
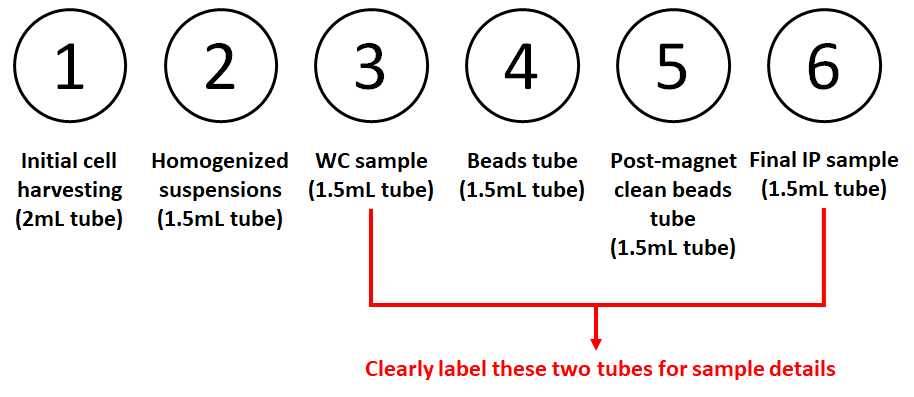
Prepare microcentrifuge tubes as follows on a metal rack on ice (for each sample, from left to right): ➀ 2 mL tube for cell suspension from harvesting; ➁ 1.5 mL tube for post-homogenization cell suspension (organelles in supernatant, membranes in pellet); ➂ 1.5 mL tube for whole cell sample; ➃ 1.5 mL tube for beads; ➄ 1.5 mL tube for post-magnetic samples; ➅ 1.5 mL tube for final Golgi-IP samples. Carefully label tubes ➂ and ➅ with detailed samples and experiments names.
Preparation of Anti-HA beads
Pool all required volumes together (100µL/ plate, e.g. 800 uL total for 8 plates, extra is not needed).
.
Shake bottle very well before removing as beads tend to sink to the bottom.
Wash 3 x with the same volume cold clean MS grade KPBS, after settling on magnet. Remove the holder from the magnet itself before dispending washing KPBS to avoid wetting the magnet.
Resuspend with KPBS with same amount of volume originally removed from bottle.
Aliquot 100µL into each 1.5 mL labeled tubes ➃.
Cell preparation before harvesting
Wash the first set of 15cm plates (each set has two plates) with 10mL of DMEM/plate (for HEKS, use no serum + no antibiotics).
Replace with 10mL of DMEM/plate for an hour. You can also use full media or other treatments based on your experimental needs.
10mL The second set of plates will be washed 0h 20m 0s later after the first set and so on.
One hour after DMEM wash, take the first set of plates from incubator to bench and place on ice.
Decant the media. Then Wash the cells twice by pouring ~5mL cold clean MS grade PBS on the edge of the plate, decant the first time and then aspirate the second time.
Cell harvesting
Add 950µL of cold KPBS to each 15-cm dish.
Scrape the cells down to the bottom of your plates with a cell lifter and transfer the cell suspension into the 2ml tube ➀. Note: this step should be carefully accounted for and done the same between plates. Visually check (with an angle) that all cells have been harvested. We are using a 2mL tube since 950 uL KPBS + cells gives around 2mL volume.
Spin at 1000x g,0h 0m 0s for 0h 2m 0s at 4°C.
Aspirate the supernatant and resuspend the pellets with 950µL cold KPBS.
From this resuspended sample, take 25µL for whole cell in the 1.5 mL tube ➂.
Note: if pellet mixer is used instead of douncer, resuspend the pellets with 100µL cold KPBS in step 16, homogenize cells and then replenish to 950 uL and follow step 17.
Homogenization and Golgi-IP
Transfer the remainder (925µL) of cells into a clean and pre-chill douncer. Dounce the cells 25 times (for HEK293T cells, other cells need to be optimized) gently on ice and avoid making bubbles.
Use 2 mL serological pipet to transfer sample from douncer into the 1.5 mL tubes ➁.
Spin 1,000g for 0h 2m 0s at 4°C.
a. Wash douncers during this spin for subsequent harvesting
Put the remaining supernatant ( it contains the organelles ) on the 1.5 ml tube ➃ with beads and resuspend by pipetting up and down ONE TIME.
Rock in cold room for 0h 3m 0s (everything from now on is in the cold room).
Put the ➃ tube on magnet. Count at least 0h 0m 25s to allow for beads to be pulled by magnets.
Wash the bound fraction 3 times with 1mLcold KPBS. Then aspirate all cold KPBS.
Processing of nonpolar lipids samples
For nonpolar metabolites (lipidomics), both IP and WC samples, resuspend in 1000µL of chloroform:methanol at ratio of 2:1 (v/v) with 1000x diluted Splashmix (Avanti). Then incubate for 0h 10m 0s .
After 0h 10m 0s of finishing the last IP, place IP samples in the tube ➄ on the magnet, collect supernatant, and transfer to the 1.5 mL tube ➅.
For both WC and Golgi-IP samples, vortex for 1h 0m 0s in cold room. Then add 200µL of 0.9% (w/v) saline (VWR) and vortex for another 0h 10m 0s in cold room. The mixture was centrifuged at 3000x g,0h 0m 0s for 0h 5m 0s at 4°C . Then discard the top layer (MeOH and saline polar phase) and use bubbeling method to retrieve 600µL from the bottom layer (chloroform containing lipids) to clean prechilled eppendorf tubes. Next speedvac the chloroform samples until dried. Then store lipidomics WC and IP samples at -80°C. On the day of analysis, dried lipid extracts were reconstituted in 50µL of ACN:IPA:water 13:6:1 (v/v/v) and vortexed for 0h 10m 0s at 4°C . Then samples are centrifuged for 0h 15m 0s at 4°C at max speed, and then 45µL of supernatant is transferred into glass insert vials for LC/MS.

